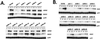Analysis of TAF90 mutants displaying allele-specific and broad defects in transcription - PubMed (original) (raw)
Analysis of TAF90 mutants displaying allele-specific and broad defects in transcription
R J Durso et al. Mol Cell Biol. 2001 Nov.
Abstract
Yeast TAF90p is a component of at least two transcription regulatory complexes, the general transcription factor TFIID and the Spt-Ada-Gcn5 histone acetyltransferase complex (SAGA). Broad transcription defects have been observed in mutants of other TAF(II)s shared by TFIID and SAGA but not in the only two TAF90 mutants isolated to date. Given that the numbers of mutants analyzed thus far are small, we isolated and characterized 11 temperature-sensitive mutants of TAF90 and analyzed their effects on transcription and integrity of the TFIID and SAGA complexes. We found that the mutants displayed a variety of allele-specific defects in their ability to support transcription and maintain the structure of the TFIID and SAGA complexes. Sequencing of the alleles revealed that all have mutations corresponding to the C terminus of the protein, with most clustering within the conserved WD40 repeats; thus, the C terminus of TAF90p is required for its incorporation into TFIID and function in SAGA. Significantly, inactivation of one allele, taf90-20, caused the dramatic reduction in the levels of total mRNA and most specific transcripts analyzed. Analysis of the structure and/or activity of both TAF90p-containing complexes revealed that this allele is the most disruptive of all. Our analysis defines the requirement for the WD40 repeats in preserving TFIID and SAGA function, demonstrates that the defects associated with distinct mutations in TAF90 vary considerably, and indicates that TAF90 can be classified as a gene required for the transcription of a large number of genes.
Figures
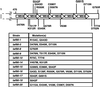
FIG. 1
Isolation of TAF90 mutants. A table of amino acid substitutions and the locations of those in the C terminus relative to the putative WD40 repeats are shown.
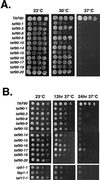
FIG. 2
Characterization of growth phenotypes. (A) Growth phenotypes on rich medium. Cultures were grown in rich medium to an OD600 of 1.0, and 2 μl of 10-fold serial dilutions was spotted onto agar plates. Plates were grown for 3 days at the temperatures indicated. (B) Recovery of TAF90 mutants from exposure to the restrictive temperature. Cultures were grown in liquid rich medium to an OD600 of 1.0, and then 2 μl of 10-fold serial dilutions was spotted onto solid rich medium. Plates were grown either for 3 days at 23°C or for 12 or 24 h at 37°C and then for 3 days at 23°C. Since the cells were plated onto agar plates at room temperature, the exact time that the cells were exposed to a temperature of 37°C is not known.
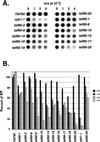
FIG. 3
Poly(A)+ RNA levels. (A) Cultures were grown in YPAD to an OD600 of approximately 0.3 and then shifted to 37°C. Aliquots were removed prior to the shift (0 h) and after 1, 2, and 4 h at 37°C, and RNA was isolated by acid-phenol extraction. Ten micrograms of total RNA was spotted onto nitrocellulose, and poly(A)+ RNA was detected by probing with a 32P-labeled dT20 probe. (B) Quantification. Spots were analyzed using a phosphorimager. The percentage of wild-type (WT) levels was determined by comparing the counts in each spot with those in the wild-type spot from 0 h. The taf90-2 strain (#) is a previously described noncogenic temperature-sensitive strain used as a control (2). The relative levels of mRNA in the mutants versus the wild type varied between 5 and 15% between experiments, but most points varied by less than 10%.
FIG. 4
Transcription analysis of specific messages. Cultures were grown, treated, and processed as described in the legend to Fig. 3. (A) S1 analysis of specific messages. Ten micrograms of total RNA was analyzed by S1 nuclease protection (7). tRNAIle, a Pol III transcript, was used as an RNA control. (B) Northern blotting. Fifteen micrograms of total RNA was fractionated on formaldehyde-containing gels and transferred to a membrane, and the specific transcripts were detected by hybridization with 32P-labeled probes corresponding to the coding regions of CLB2, CLN2, TRX1, and RPS5. The Pol III-transcribed scR1 was used as a loading control.
FIG. 5
Induced transcription. (A) Derepression of SUC2 and ADH2. Cultures of YJR241-x were grown initially in YPAD at 23°C. Following the removal of an aliquot from each culture, cells were centrifuged, resuspended in YPA containing a low concentration of dextrose (0.05%) prewarmed to 37°C, and grown at the restrictive temperature. RNA was isolated from aliquots that were removed after 1.5 and 3 h at 37°C in low-dextrose medium. Transcripts were detected by Northern blotting. scR1 was used as a loading control. (B) Analysis of a DNA damage-induced gene. Cultures of YJR241-x were grown in rich medium and shifted to the nonpermissive temperature. Aliquots were removed (permissive [lanes P]) prior to the temperature shift. After 1 h at 37°C, MMS was added to 0.02%. Aliquots were taken at 1.5 and 3 h after treatment with MMS. A separate culture was maintained at 37°C but left untreated (nonpermissive [lanes N]) for 4 h. HUG1 RNA was analyzed by Northern blotting. scR1 was used as a loading control.
FIG. 6
Effects of TAF90 mutations on Gcn4p-mediated transcription. Cells were grown under noninducing conditions in synthetic complete medium at 24°C (lanes N) and at 37°C for 3 h (lanes N, 37). A separate culture was grown in synthetic complete medium without histidine, shifted to 37°C for 1.5 h, and incubated for another 1.5 h in the presence of 30 mM 3-aminotriazole (lanes I, 37). Analysis of HIS3 was performed by S1 nuclease protection. The induction level (+13 I/N) is the ratio of the +13 transcript from cells grown under inducing conditions at 37°C to that from cells grown under noninducing conditions at 37°C. The reduced tRNA signal in the taf90-19 sample is a gel-loading artifact and is not reproducible. WT, wild type.
FIG. 7
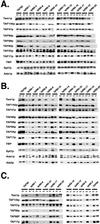
Steady-state levels of TFIID and SAGA subunits following a temperature shift. Wild-type and mutant strains were grown at 24°C in YPAD to an OD600 of approximately 0.4, and after removal of an aliquot for the t = 0 time point, they were transferred to a 37°C shaking water bath. Aliquots of culture were removed after 1, 2, and 4 h at the nonpermissive temperature. Extracts prepared from these cells (20 μg of protein) were fractionated by SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose, and the specific proteins were detected by immunoblotting.
FIG. 8
Co-IP studies with extracts of TAF90 mutants. Whole-cell extracts were prepared from cells grown at room temperature (approximately 24°C) or at 37°C for 3 h. Complexes containing TAF90p were immunoprecipitated from cell extracts using antibodies raised against TAF90p or TAF145p. Cell extracts and IPs were analyzed by Western blotting using the antibodies indicated. The input material (A) and complexes immunoprecipitated using antibody raised against TAF90p (B) or TAF145p (C) are shown.
FIG. 9
Purification of HAT complexes from TAF90 mutants. Extracts prepared from mutants grown at 37°C for 3 h were subjected to chromatographic analysis as described in Materials and Methods. Aliquots of the Mono Q column fractions were analyzed by Western blotting and HAT assays using nucleosomes as substrates. Elution profiles for the wild type (A) and the taf90-20 (B), taf90-17 (C), and taf90-10 (D) mutants are shown. Note that the blots presented in panel D were deliberately exposed longer than those in panels A to C to detect the small amounts of SAGA subunits eluting in fractions 38 to 42.
FIG. 10
Analysis of mutant SAGA complexes. (A) SAGA has reduced HAT activity in TAF90 mutants. Fractions containing SAGA (fractions 38 to 42 from Fig. 9) were pooled and analyzed by Western blotting and HAT assays using oligonucleosomes as substrates. The relative levels of Gcn5p were determined from appropriately exposed blots using NIH Image software and are expressed as the percentage of the wild-type (WT) level. HAT activity was compared to that of SAGA isolated from wild-type cells, which was set at 100%. (B) Size exclusion chromatography. The pooled SAGA-containing fractions from the Mono Q chromatography (fractions 38 to 42 from Fig. 9) were concentrated by filtration and fractionated on a Superose 6 column. Each fraction was analyzed by Western blotting and HAT assays using nucleosomes as a substrate. The void volume corresponds to fraction 14. No HAT activity or SAGA components were detected in later-eluting fractions, and thus only fractions 15 to 23 are shown.
Similar articles
- Distinct mutations in yeast TAF(II)25 differentially affect the composition of TFIID and SAGA complexes as well as global gene expression patterns.
Kirschner DB, vom Baur E, Thibault C, Sanders SL, Gangloff YG, Davidson I, Weil PA, Tora L. Kirschner DB, et al. Mol Cell Biol. 2002 May;22(9):3178-93. doi: 10.1128/MCB.22.9.3178-3193.2002. Mol Cell Biol. 2002. PMID: 11940675 Free PMC article. - Redundant roles for the TFIID and SAGA complexes in global transcription.
Lee TI, Causton HC, Holstege FC, Shen WC, Hannett N, Jennings EG, Winston F, Green MR, Young RA. Lee TI, et al. Nature. 2000 Jun 8;405(6787):701-4. doi: 10.1038/35015104. Nature. 2000. PMID: 10864329 - Adenovirus E1A requires the yeast SAGA histone acetyltransferase complex and associates with SAGA components Gcn5 and Tra1.
Kulesza CA, Van Buskirk HA, Cole MD, Reese JC, Smith MM, Engel DA. Kulesza CA, et al. Oncogene. 2002 Feb 21;21(9):1411-22. doi: 10.1038/sj.onc.1205201. Oncogene. 2002. PMID: 11857084 - SAGA and TFIID: Friends of TBP drifting apart.
Timmers HTM. Timmers HTM. Biochim Biophys Acta Gene Regul Mech. 2021 Feb;1864(2):194604. doi: 10.1016/j.bbagrm.2020.194604. Epub 2020 Jul 14. Biochim Biophys Acta Gene Regul Mech. 2021. PMID: 32673655 Review. - Recruitment of chromatin remodelling factors during gene activation via the glucocorticoid receptor N-terminal domain.
Wallberg AE, Flinn EM, Gustafsson JA, Wright AP. Wallberg AE, et al. Biochem Soc Trans. 2000;28(4):410-4. Biochem Soc Trans. 2000. PMID: 10961930 Review.
Cited by
- Spt-Ada-Gcn5-Acetyltransferase (SAGA) Complex in Plants: Genome Wide Identification, Evolutionary Conservation and Functional Determination.
Srivastava R, Rai KM, Pandey B, Singh SP, Sawant SV. Srivastava R, et al. PLoS One. 2015 Aug 11;10(8):e0134709. doi: 10.1371/journal.pone.0134709. eCollection 2015. PLoS One. 2015. PMID: 26263547 Free PMC article. - Mapping key functional sites within yeast TFIID.
Leurent C, Sanders SL, Demény MA, Garbett KA, Ruhlmann C, Weil PA, Tora L, Schultz P. Leurent C, et al. EMBO J. 2004 Feb 25;23(4):719-27. doi: 10.1038/sj.emboj.7600111. Epub 2004 Feb 12. EMBO J. 2004. PMID: 14765106 Free PMC article. - TFIID dependency of steady-state mRNA transcription altered epigenetically by simultaneous functional loss of Taf1 and Spt3 is Hsp104-dependent.
Iwami R, Takai N, Matsutani M, Shiwa Y, Kokubo H, Kasahara K, Kokubo T. Iwami R, et al. PLoS One. 2023 Feb 9;18(2):e0281233. doi: 10.1371/journal.pone.0281233. eCollection 2023. PLoS One. 2023. PMID: 36757926 Free PMC article. - Distinct mutations in yeast TAF(II)25 differentially affect the composition of TFIID and SAGA complexes as well as global gene expression patterns.
Kirschner DB, vom Baur E, Thibault C, Sanders SL, Gangloff YG, Davidson I, Weil PA, Tora L. Kirschner DB, et al. Mol Cell Biol. 2002 May;22(9):3178-93. doi: 10.1128/MCB.22.9.3178-3193.2002. Mol Cell Biol. 2002. PMID: 11940675 Free PMC article. - Dissection of coactivator requirement at RNR3 reveals unexpected contributions from TFIID and SAGA.
Zhang H, Kruk JA, Reese JC. Zhang H, et al. J Biol Chem. 2008 Oct 10;283(41):27360-27368. doi: 10.1074/jbc.M803831200. Epub 2008 Aug 5. J Biol Chem. 2008. PMID: 18682387 Free PMC article.
References
- Albright S R, Tjian R. TAFs revisited: more data reveal new twists and confirm old ideas. Gene. 2000;242:1–13. - PubMed
- Apone L M, Virbasius C M, Reese J C, Green M R. Yeast TAF(II)90 is required for cell-cycle progression through G2/M but not for general transcription activation. Genes Dev. 1996;10:2368–2380. - PubMed
- Apone L M, Virbasius C A, Holstege F C, Wang J, Young R A, Green M R. Broad, but not universal, transcriptional requirement for yTAFII17, a histone H3-like TAFII present in TFIID and SAGA. Mol Cell. 1998;2:653–661. - PubMed
- Brachmann C B, Davies A, Cost G J, Caputo E, Li J, Heiter P, Boeke J D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials