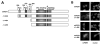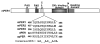Nuclear export of mammalian PERIOD proteins - PubMed (original) (raw)
Nuclear export of mammalian PERIOD proteins
E L Vielhaber et al. J Biol Chem. 2001.
Abstract
The timing of mammalian circadian rhythm is determined by interlocking negative and positive transcriptional feedback loops that govern the cyclic expression of both clock regulators and output genes. In mammals, nuclear localization of the circadian regulators PER1-3 is controlled by multiple mechanisms, including multimerization with PER and CRY proteins. In addition, nuclear entry of mammalian PER1 (mPER1) can be regulated by a phosphorylation-dependent masking of its nuclear localization signal. Here we present evidence suggesting that nuclear localization of PER proteins is a dynamic process determined by both nuclear import and previously unrecognized nuclear export pathways. Examination of the subcellular localization of a series of truncated mPER1 proteins demonstrated that cytoplasmic localization is mediated by an 11-amino acid region with homology to leucine-rich nuclear export signals (NESs). Similar sequences were identified in mPER2 and mPER3 as well as in several insect PER proteins. The putative NESs from mPER1 and mPER2 were able to direct cytoplasmic accumulation when fused to a heterologous protein. Mutations in conserved NES residues and the nuclear export inhibitor leptomycin B each blocked the function of the NES. Full-length mPER1 was also exported from microinjected Xenopus laevis oocyte nuclei in an NES-dependent manner. The presence of a functional NES in mPER1 and mPER2 as well as related sequences in a variety of other PER proteins suggests that nuclear export may be a conserved and important feature of circadian regulators.
Figures
Fig. 1. Successive amino-terminal truncations of mPER1 reveal that two distinct regions are important for mediating the subcellular localization of mPER1
A, diagram of the mPER1 truncation mutants. The truncated forms of mPER1 used in B are drawn, with previously described domains within mPER1 indicated by boxes in varying shades of gray and the putative NES indicated by hatched boxes. The PAS A and B domains and CLD (10, 28) and the NLS, casein kinase Iε (_CKI_ε)-binding region, and NLS-masking domain (16) have been described. B, the amino-terminal region of mPER1 (amino acids 1–205) is important for the nuclear localization of mPER1, whereas amino acids 485–495 are important for cytoplasmic localization. HEK 293 cells were transfected with the indicated Myc-mPER1 constructs, and the subcellular location of the proteins was determined with immunofluorescence. The Myc-tagged mPER1 proteins were detected with an anti-Myc monoclonal antibody directly coupled to Alexa 488, and nuclei were visualized by Hoechst staining. Representative micrographs are shown. The subcellular localization of the various mPER proteins depicted in the micrographs was observed in >95% of the cells. Experiments were repeated at least twice, and >100 cells were counted for each transfected construct.
Fig. 2. Numerous PER proteins contain a putative NES carboxyl-terminal to the CLD
The positions and sequences of the putative NES regions from mPER1, mPER2, mPER3, dPER, and A. pernyi (ap) PER are shown in relation to a diagram of the full-length mPER1 protein. The hydrophobic residues known to be important for NES function in other proteins are shown in boldface. The PER sequences are aligned based on the position of these hydrophobic residues. Numbers indicate the locations of the amino acids within the respective proteins. For comparison, a consensus sequence of the leucine-rich NES is shown (29). Although originally defined as “leucine-rich,” other hydrophobic residues (Ile, Phe, Val, and Met) have been shown to substitute for the second through fourth leucines in functional NES sequences of various proteins (26, 29). Within the consensus sequence, X denotes any amino acid. _CKI_ε, casein kinase Iε.
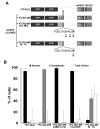
Fig. 3. Fusion of the CLD/NES region of mPER1 (amino acids 411–498) to a YFP carrier protein promotes nuclear export
A, diagrams of the YFP carrier protein (YFP-NLS) and the YFP-CLD/NES fusion proteins. The YFP fusion proteins used in B–D are drawn below a full-length mPER1 diagram, and the specific mutations made in the NES are indicated below the NES sequence. The regions of mPER1 fused to the YFP-NLS protein are indicated by +P1 plus the corresponding amino acid numbers. All regions of mPER1 fused to YFP are aligned with the respective domains of full-length mPER1. B, the mPER1 NES alone controls the cytoplasmic localization of YFP. HEK 293 cells were transfected with the indicated Myc-mPER1 constructs, and the subcellular location of the proteins was determined by autofluorescence of YFP. Nuclei were visualized by Hoechst staining. Representative micrographs are shown. C, the cytoplasmic localization of the YFP-CLD/NES fusion proteins can be inhibited by LMB. HEK 293 cells were transfected with the indicated YFP fusion constructs and treated with vehicle (*minus; LMB) or with LMB (+ LMB) for 4 h. Following the treatment, the subcellular location of the proteins was determined by autofluorescence of YFP, and nuclei were visualized by Hoechst staining. Representative micrographs are shown. D, quantitation of the experiments illustrated above. Black bars represent predominantly nuclear localization; gray bars represent predominantly cytoplasmic localization; and white bars represent equal localization in the cytoplasm and nucleus. All experiments were performed at least twice, and >100 cells were counted for each condition tested. The lack of error bars for the LMB data indicates that replicate experiments produced identical results.
Fig. 4. Fusion of the CLD/NES region of mPER2 (amino acids 382–469) to a YFP carrier protein promotes nuclear export
A, diagrams of the YFP carrier protein (YFP-NLS) and the YFP-CLD/NES fusion proteins. The YFP fusion proteins used in B are drawn. + P2 indicates the mPER2 protein and is followed by the corresponding amino acid numbers used in the fusion with YFP-NLS. The specific mutations made in the NES are indicated below each NES sequence. The region of mPER1 fused to YFP to promote nuclear localization is indicated above that region. B, fusion of the CLD/NES region of mPER2 to YFP promotes the cytoplasmic localization of YFP and can be inhibited by LMB or mutation of the NES sequence. HEK 293 cells were transfected with the indicated YFP fusion constructs, and one set of cells expressing the +P2-(382–469) construct was treated with LMB for 4 h. The subcellular location of the proteins was determined by autofluorescence of YFP, and nuclei were visualized by Hoechst staining. Black bars represent predominantly nuclear localization; gray bars represent predominantly cytoplasmic localization; and white bars represent equal localization in the cytoplasm and nucleus. All experiments were performed at least twice, and >100 cells were counted for each condition tested.
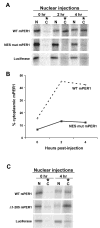
Fig. 5. mPER1 is exported from the nuclei of X. laevis oocytes
A, nuclear export of mPER1 in X. laevis oocytes requires a functional NES. The indicated 35S-labeled proteins were injected into the nuclei of X. laevis oocytes. At the indicated times following injection, the nuclei were separated from the cytoplasm, and the nuclear and cytoplasmic fractions were analyzed separately by SDS-PAGE. Each lane contains the proteins from the nucleus or cytoplasm of one oocyte, respectively. In all experiments, luciferase was co-injected with wild-type (WT mPER1) or mutant mPER1 (NES mut mPER1), but only the luciferase co-injected with wt mPER1 is shown. The NES mutant mPER1 protein has Ile-493 and Leu-496 mutated to alanine. These mutations are in the context of the full-length mPER1 protein. This experiment was repeated several times with similar results. B, quantitation of the amount of mPER1 protein found in the cytoplasm at various times post-injection. The amount of mPER1 protein in the oocyte cytoplasm at the indicated times following nuclear injection is expressed as a percentage of the total amount of mPER1 in the oocyte (nuclear fraction + cytoplasmic fraction) at each time point. C, the rate of cytoplasmic accumulation is controlled in part by the amino terminus of mPER1. The indicated 35S-labeled proteins were injected into the nuclei of X. laevis oocytes, and the experiment was carried out as described for A. The Δ 1–205 mPER1 protein is the same as the protein shown in Fig. 1_A_. This experiment was repeated several times with similar results. N, nuclear fraction; C, cytoplasmic fraction.
Similar articles
- Nuclear import of mPER3 in Xenopus oocytes and HeLa cells requires complex formation with mPER1.
Loop S, Pieler T. Loop S, et al. FEBS J. 2005 Jul;272(14):3714-24. doi: 10.1111/j.1742-4658.2005.04798.x. FEBS J. 2005. PMID: 16008569 - Nucleocytoplasmic shuttling of clock proteins.
Tamanini F, Yagita K, Okamura H, van der Horst GT. Tamanini F, et al. Methods Enzymol. 2005;393:418-35. doi: 10.1016/S0076-6879(05)93020-6. Methods Enzymol. 2005. PMID: 15817303 - Nuclear entry of the circadian regulator mPER1 is controlled by mammalian casein kinase I epsilon.
Vielhaber E, Eide E, Rivers A, Gao ZH, Virshup DM. Vielhaber E, et al. Mol Cell Biol. 2000 Jul;20(13):4888-99. doi: 10.1128/MCB.20.13.4888-4899.2000. Mol Cell Biol. 2000. PMID: 10848614 Free PMC article. - mPER1-mediated nuclear export of mCRY1/2 is an important element in establishing circadian rhythm.
Loop S, Katzer M, Pieler T. Loop S, et al. EMBO Rep. 2005 Apr;6(4):341-7. doi: 10.1038/sj.embor.7400372. EMBO Rep. 2005. PMID: 15791269 Free PMC article. - Leucine-rich nuclear-export signals: born to be weak.
Kutay U, Güttinger S. Kutay U, et al. Trends Cell Biol. 2005 Mar;15(3):121-4. doi: 10.1016/j.tcb.2005.01.005. Trends Cell Biol. 2005. PMID: 15752974 Review.
Cited by
- Casein kinase I in the mammalian circadian clock.
Eide EJ, Kang H, Crapo S, Gallego M, Virshup DM. Eide EJ, et al. Methods Enzymol. 2005;393:408-18. doi: 10.1016/S0076-6879(05)93019-X. Methods Enzymol. 2005. PMID: 15817302 Free PMC article. - Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS).
Vanselow K, Vanselow JT, Westermark PO, Reischl S, Maier B, Korte T, Herrmann A, Herzel H, Schlosser A, Kramer A. Vanselow K, et al. Genes Dev. 2006 Oct 1;20(19):2660-72. doi: 10.1101/gad.397006. Epub 2006 Sep 18. Genes Dev. 2006. PMID: 16983144 Free PMC article. - Of switches and hourglasses: regulation of subcellular traffic in circadian clocks by phosphorylation.
Tataroğlu O, Schafmeier T. Tataroğlu O, et al. EMBO Rep. 2010 Dec;11(12):927-35. doi: 10.1038/embor.2010.174. Epub 2010 Nov 5. EMBO Rep. 2010. PMID: 21052092 Free PMC article. - Setting clock speed in mammals: the CK1 epsilon tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins.
Meng QJ, Logunova L, Maywood ES, Gallego M, Lebiecki J, Brown TM, Sládek M, Semikhodskii AS, Glossop NRJ, Piggins HD, Chesham JE, Bechtold DA, Yoo SH, Takahashi JS, Virshup DM, Boot-Handford RP, Hastings MH, Loudon ASI. Meng QJ, et al. Neuron. 2008 Apr 10;58(1):78-88. doi: 10.1016/j.neuron.2008.01.019. Neuron. 2008. PMID: 18400165 Free PMC article. - Molecular Aspects of Circadian Pharmacology and Relevance for Cancer Chronotherapy.
Ozturk N, Ozturk D, Kavakli IH, Okyar A. Ozturk N, et al. Int J Mol Sci. 2017 Oct 17;18(10):2168. doi: 10.3390/ijms18102168. Int J Mol Sci. 2017. PMID: 29039812 Free PMC article. Review.
References
- Dunlap JC. Cell. 1999;96:271–290. - PubMed
- Reppert SM, Weaver DR. Annu Rev Physiol. 2001;63:647–676. - PubMed
- Lowrey PL, Takahashi JS. Annu Rev Genet. 2000;34:533–562. - PubMed
- Curtin KD, Huang ZJ, Rosbash M. Neuron. 1995;14:365–372. - PubMed
- Ponting CP, Aravind L. Curr Biol. 1997;7:R674–R677. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases