The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins - PubMed (original) (raw)
The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins
T C Walther et al. EMBO J. 2001.
Abstract
The nuclear pore complex (NPC) is a large proteinaceous structure through which bidirectional transport of macromolecules across the nuclear envelope (NE) takes place. Nup153 is a peripheral NPC component that has been implicated in protein and RNP transport and in the interaction of NPCs with the nuclear lamina. Here, Nup153 is localized by immunogold electron microscopy to a position on the nuclear ring of the NPC. Nuclear reconstitution is used to investigate the role of Nup153 in nucleo- cytoplasmic transport and NPC architecture. NPCs assembled in the absence of Nup153 lacked several nuclear basket components, were unevenly distributed in the NE and, unlike wild-type NPCs, were mobile within the NE. Importin alpha/beta-mediated protein import into the nucleus was strongly reduced in the absence of Nup153, while transportin-mediated import was unaffected. This was due to a reduction in import complex translocation rather than to defective receptor recycling. Our results therefore reveal functions for Nup153 in NPC assembly, in anchoring NPCs within the NE and in mediating specific nuclear import events.
Figures
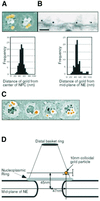
Fig. 1. Localization of Nup153 within the NPC. Fixed Xenopus oocyte nuclear envelopes were incubated with affinity-purified anti-Nup153 primary antibody and 10 nm colloidal gold labelled secondary antibodies and analysed using electron microscopy. (A) Upper panel: representative FEISEM image of NPCs decorated with gold particles (false coloured yellow). Gold particles were identified using the backscatter electron image. Lower panel: distribution of distances to the centre of the NPC (in nm) of 100 NPC-associated gold particles. Bar, 100 nm. (B) Upper panel: representative TEM image of a stretch of nuclear envelope (NE) decorated with anti-Nup153 primary antibodies and gold labelled secondary antibodies. Bar, 100 nm. Lower panel: distribution of distances to the mid-plane of the NE (in nm) of 85 NE-associated gold particles. (C) Representative images of immunogold localization of Nup153 on NPCs after stripping of the nucleoplasmic basket. Bar, 100 nm. (D) Schematic representation of the localization of Nup153-specific gold labelling on a diagram of the NPC basket. Note that two antibody molecules (not shown in the diagram) are interposed between Nup153 and the gold particle. Bars indicate standard deviations of measurement.
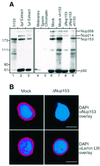
Fig. 2. Generation of Nup153-deficient nuclei. (A) Western blot probed with mAb414. Left panel: 1 µl of recombinant hNup153 (lane 1) compared with 2 and 1 µl of undepleted Xenopus egg cytosol fraction (lanes 2 and 3). Right panel: 2 µl of Xenopus egg membrane fraction, 2 µl of sperm chromatin, 1 µl of either mock or Nup153-depleted nuclear assembly reaction with or without addition of recombinant human Nup153, as indicated above the lanes. Molecular weight markers (kDa) are shown on the left. Positions of mAb414-reactive nucleoporins are shown on the right. (B) Immunofluorescence staining of nuclei formed in the mock-depleted or the Nup153-depleted (ΔNup153) extracts using monospecific anti-Nup153 antibodies or monoclonal S49 against lamin LIII (Lourim and Krohne, 1993, 1998). As secondary antibodies, Alexa-546 conjugates were used (red). Decondensed sperm chromatin is stained with DAPI (blue). Bar, 10 µm.
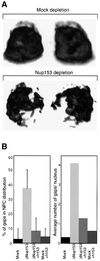
Fig. 3. Clustering of Nup153-deficient NPCs. Nuclei were assembled in vitro from mock-depleted or Nup153-depleted Xenopus egg cytosol and NPC distribution analysed using confocal immunofluorescence. (A) 3D reconstruction of confocal sections through a wild-type (Mock depletion) and Nup153-deficient (Nup153 depletion) nucleus stained with anti-Nup214/CAN primary and Alexa-488 conjugated secondary antibodies. Left hand images are rotated 180° as compared with right hand images. (B) Quantitation of NPC clustering. Nuclei were assembled from mock or Nup153-depleted (ΔNup153) Xenopus egg extracts in the presence (+h153) or absence of recombinant hNup153, and immunostained with mAb414 and Alexa-488 conjugated secondary antibody. Mid-plane confocal images were recorded and NEs were analysed for discontinuities. Both the percentage of NE length without nucleoporin staining (left panel) and the number of nucleoporin gaps in the nucleoporin staining per nucleus (right panel) are represented. Bars represent standard deviations. (C) Representative immunofluorescence images of mid-plane confocal sections of synthetic nuclei analysed in (B). Nucleoporin staining is in red, chromatin staining (DAPI) in blue.
Fig. 3. Clustering of Nup153-deficient NPCs. Nuclei were assembled in vitro from mock-depleted or Nup153-depleted Xenopus egg cytosol and NPC distribution analysed using confocal immunofluorescence. (A) 3D reconstruction of confocal sections through a wild-type (Mock depletion) and Nup153-deficient (Nup153 depletion) nucleus stained with anti-Nup214/CAN primary and Alexa-488 conjugated secondary antibodies. Left hand images are rotated 180° as compared with right hand images. (B) Quantitation of NPC clustering. Nuclei were assembled from mock or Nup153-depleted (ΔNup153) Xenopus egg extracts in the presence (+h153) or absence of recombinant hNup153, and immunostained with mAb414 and Alexa-488 conjugated secondary antibody. Mid-plane confocal images were recorded and NEs were analysed for discontinuities. Both the percentage of NE length without nucleoporin staining (left panel) and the number of nucleoporin gaps in the nucleoporin staining per nucleus (right panel) are represented. Bars represent standard deviations. (C) Representative immunofluorescence images of mid-plane confocal sections of synthetic nuclei analysed in (B). Nucleoporin staining is in red, chromatin staining (DAPI) in blue.
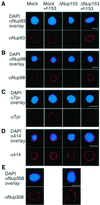
Fig. 4. Nup153-deficient NPCs lack several other components. Nuclei were assembled in vitro from mock-depleted (Mock) or Nup153-depleted (ΔNup153) Xenopus egg cytosol in the presence (+h153) or absence of recombinant hNup153, as indicated. Assembled nuclei were fixed and immunostained with antibodies recognizing Nup93 (A), Nup98 (B), Tpr (C), with mAb414 (D) or Nup358/RanBP2 (E). Secondary antibodies were conjugated to Alexa-546 (red). Decondensed sperm chromatin is stained with DAPI (blue). Bar, 10 µm.
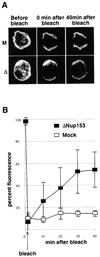
Fig. 5. Nup153-deficient NPCs display increased mobility within the NE. (A) Representative fluorescence recovery after photobleaching (FRAP) series of nuclei assembled in vitro from mock-depleted (M, upper panels) or Nup153-depleted (Δ, lower panels) Xenopus egg extracts, stained with Oregon Orange labelled wheatgerm agglutinin. (B) Quantitation of FRAP in three independent experiments using mock-depleted (open squares) or Nup153-depleted (solid squares) assembled nuclei.
Fig. 6. Nup153-deficient NPCs display a selective protein import defect. Nuclei were assembled in vitro from mock-depleted (Mock) or Nup153-depleted (ΔNup153) Xenopus egg cytosol in the presence (+h153) or absence of recombinant hNup153 as indicated. (A) Quantitation of import of fluorescently labelled M9 (left panel) or classical NLS (right panel) import substrates in randomly selected nuclei from different experiments. Import is represented as the percentage of the nuclear fluorescence in mock-depleted nuclei. Bars show standard deviations. (B) Representative images of fluorescently labelled BSA–NLS import into nuclei as analysed in (A). Note the lack of nuclear rim staining in the bottom left panel. (C) Quantitation of nuclear importin α and importin β by immunofluorescence and direct fluorescence measurement. (D) Representative images of nuclei stained for importin α (α) or importin β (β), as quantified in (C).
Similar articles
- Nuclear protein import is reduced in cells expressing nuclear envelopathy-causing lamin A mutants.
Busch A, Kiel T, Heupel WM, Wehnert M, Hübner S. Busch A, et al. Exp Cell Res. 2009 Aug 15;315(14):2373-85. doi: 10.1016/j.yexcr.2009.05.003. Epub 2009 May 12. Exp Cell Res. 2009. PMID: 19442658 - Recombinant Nup153 incorporates in vivo into Xenopus oocyte nuclear pore complexes.
Panté N, Thomas F, Aebi U, Burke B, Bastos R. Panté N, et al. J Struct Biol. 2000 Apr;129(2-3):306-12. doi: 10.1006/jsbi.2000.4232. J Struct Biol. 2000. PMID: 10806081 - Mediators of nuclear protein import target karyophilic proteins to pore complexes of cytoplasmic annulate lamellae.
Cordes VC, Rackwitz HR, Reidenbach S. Cordes VC, et al. Exp Cell Res. 1997 Dec 15;237(2):419-33. doi: 10.1006/excr.1997.3806. Exp Cell Res. 1997. PMID: 9434638 - Function and assembly of nuclear pore complex proteins.
Bodoor K, Shaikh S, Enarson P, Chowdhury S, Salina D, Raharjo WH, Burke B. Bodoor K, et al. Biochem Cell Biol. 1999;77(4):321-9. Biochem Cell Biol. 1999. PMID: 10546895 Review. - Dynamic nuclear pore complexes: life on the edge.
Tran EJ, Wente SR. Tran EJ, et al. Cell. 2006 Jun 16;125(6):1041-53. doi: 10.1016/j.cell.2006.05.027. Cell. 2006. PMID: 16777596 Review.
Cited by
- The Nup153-Nup50 protein interface and its role in nuclear import.
Makise M, Mackay DR, Elgort S, Shankaran SS, Adam SA, Ullman KS. Makise M, et al. J Biol Chem. 2012 Nov 9;287(46):38515-22. doi: 10.1074/jbc.M112.378893. Epub 2012 Sep 24. J Biol Chem. 2012. PMID: 23007389 Free PMC article. - Types of nuclear localization signals and mechanisms of protein import into the nucleus.
Lu J, Wu T, Zhang B, Liu S, Song W, Qiao J, Ruan H. Lu J, et al. Cell Commun Signal. 2021 May 22;19(1):60. doi: 10.1186/s12964-021-00741-y. Cell Commun Signal. 2021. PMID: 34022911 Free PMC article. Review. - Nuclear envelope breakdown in starfish oocytes proceeds by partial NPC disassembly followed by a rapidly spreading fenestration of nuclear membranes.
Lénárt P, Rabut G, Daigle N, Hand AR, Terasaki M, Ellenberg J. Lénárt P, et al. J Cell Biol. 2003 Mar 31;160(7):1055-68. doi: 10.1083/jcb.200211076. Epub 2003 Mar 24. J Cell Biol. 2003. PMID: 12654902 Free PMC article. - Inhibition of nuclear import by the proapoptotic protein CC3.
King FW, Shtivelman E. King FW, et al. Mol Cell Biol. 2004 Aug;24(16):7091-101. doi: 10.1128/MCB.24.16.7091-7101.2004. Mol Cell Biol. 2004. PMID: 15282309 Free PMC article. - Dunking into the Lipid Bilayer: How Direct Membrane Binding of Nucleoporins Can Contribute to Nuclear Pore Complex Structure and Assembly.
Hamed M, Antonin W. Hamed M, et al. Cells. 2021 Dec 20;10(12):3601. doi: 10.3390/cells10123601. Cells. 2021. PMID: 34944108 Free PMC article. Review.
References
- Allen T.D., Bennion,G.R., Rutherpord,S.A., Reipert,S., Ramalho,A., Kiseleva,E. and Goldberg,M.W. (1997) Macromolecular substructure in nuclear pore complexes by in-lens field-emission scanning electron microscopy. Scanning, 19, 403–410. - PubMed
- Arts G.J., Englmeier,L. and Mattaj,I.W. (1997) Energy- and temperature-dependent in vitro export of RNA from synthetic nuclei. Biol. Chem., 378, 641–649. - PubMed
- Bayliss R., Littlewood,T. and Stewart,M. (2000) Structural basis for the interaction between FxFG nucleoporin repeats and importin-β in nuclear trafficking. Cell, 102, 99–108. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases