Role of Streptococcus gordonii amylase-binding protein A in adhesion to hydroxyapatite, starch metabolism, and biofilm formation - PubMed (original) (raw)
Role of Streptococcus gordonii amylase-binding protein A in adhesion to hydroxyapatite, starch metabolism, and biofilm formation
J D Rogers et al. Infect Immun. 2001 Nov.
Abstract
Interactions between bacteria and salivary components are thought to be important in the establishment and ecology of the oral microflora. alpha-Amylase, the predominant salivary enzyme in humans, binds to Streptococcus gordonii, a primary colonizer of the tooth. Previous studies have implicated this interaction in adhesion of the bacteria to salivary pellicles, catabolism of dietary starches, and biofilm formation. Amylase binding is mediated at least in part by the amylase-binding protein A (AbpA). To study the function of this protein, an erythromycin resistance determinant [erm(AM)] was inserted within the abpA gene of S. gordonii strains Challis and FAS4 by allelic exchange, resulting in abpA mutant strains Challis-E1 and FAS4-E1. Comparison of the wild-type and mutant strains did not reveal any significant differences in colony morphology, biochemical metabolic profiles, growth in complex or defined media, surface hydrophobicity, or coaggregation properties. Scatchard analysis of adhesion isotherms demonstrated that the wild-type strains adhered better to human parotid-saliva- and amylase-coated hydroxyapatite than did the AbpA mutants. In contrast, the mutant strains bound to whole-saliva-coated hydroxyapatite to a greater extent than did the wild-type strains. While the wild-type strains preincubated with purified salivary amylase grew well in defined medium with potato starch as the sole carbohydrate source, the AbpA mutants did not grow under the same conditions even after preincubation with amylase. In addition, the wild-type strain produced large microcolonies in a flow cell biofilm model, while the abpA mutant strains grew much more poorly and produced relatively small microcolonies. Taken together, these results suggest that AbpA of S. gordonii functions as an adhesin to amylase-coated hydroxyapatite, in salivary-amylase-mediated catabolism of dietary starches and in human saliva-supported biofilm formation by S. gordonii.
Figures
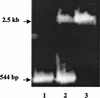
FIG. 1
PCR screening of abpA::erm(AM) recombinants of S. gordonii. Lane 1, abpA (544-bp) positive control amplified from Challis; lane 2, recombinant in S. gordonii Challis-E2 resulting from the duplication (single crossover) of abpA (544 bp) and insertion of abpA::erm(AM) (2.5 kb); lane 3, recombinant in S. gordonii Challis-E1 resulting from allelic exchange (double crossover) of abpA::erm(AM) (2.5-kb band only).
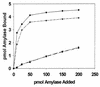
FIG. 2
Binding of 125I-labeled amylase to S. gordonii strains Challis (●), Challis-E1 (○), FAS4 (▾), FAS4-E1 (▿), and S. sanguis 10556 (▪). Data points represent the mean values of duplicate samples from two experiments.
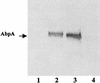
FIG. 3
Comparison of culture supernatant levels of AbpA from wild-type and AbpA mutants of S. gordonii. An amylase ligand-binding assay was used. Lanes are loaded with equal amounts of protein (25 μg) from culture supernatants of S. gordonii Challis-E1 (lane 1), Challis (lane 2), FAS4 (lane 3), and FAS4-E1 (lane 4).
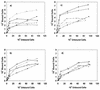
FIG. 4
Binding isotherms for the adherence of S. gordonii strains to coated HAP. (a) Whole-saliva-coated HAP; (b) HPS-coated HAP; (c) glycosylated amylase-coated HAP; (d) nonglycosylated amylase-coated HAP. Data for the following S. gordonii strains are shown: Challis (●), Challis-E1 (○), FAS4 (▾), FAS4-E1 (▿), and S. sanguis 10556 (▪).
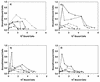
FIG. 5
Scatchard plots for adherence of S. gordonii strains to coated HAP. (a) Whole-saliva-coated HAP; (b) HPS-coated HAP; (c) glycosylated amylase-coated HAP; (d) nonglycosylated amylase-coated HAP. Data for the following S. gordonii strains are shown: Challis (●), Challis-E1 (○), FAS4 (▾), FAS4-E1 (▿), and S. sanguis 10556 (▪).
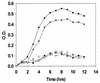
FIG. 6
Amylase-facilitated growth of amylase-binding and nonbinding streptococcal strains. Bacteria were grown in a defined medium with 2% potato starch as the primary carbon source, and growth was monitored spectrophotometrically at 600 nm. All cells were incubated with amylase prior to inoculation into the culture medium. Data for the following S. gordonii strains are shown: Challis (●), Challis-E1 (○), FAS4 (▾), FAS4-E1 (▿), and S. sanguis 10556 (▪). Data represent mean values of three experiments, each done in duplicate.
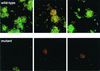
FIG. 7
Live/Dead-stained biofilms of three random fields of view from the flow cell containing S. gordonii Challis (top row) and three random fields of view of the abpA mutant Challis-E1 (bottom row) after overnight growth on 25% saliva. Image dimensions are 250 by 250 μm. Maximum microcolony height is about 10 μm (wild type) and about 5 μm (mutant). The vast majority of cells stained green in the wild-type biofilm, but the majority of the cells were red to orange in the mutant biofilm.
Similar articles
- Amylase-binding proteins A (AbpA) and B (AbpB) differentially affect colonization of rats' teeth by Streptococcus gordonii.
Tanzer JM, Grant L, Thompson A, Li L, Rogers JD, Haase EM, Scannapieco FA. Tanzer JM, et al. Microbiology (Reading). 2003 Sep;149(Pt 9):2653-2660. doi: 10.1099/mic.0.26022-0. Microbiology (Reading). 2003. PMID: 12949189 - Interaction of salivary alpha-amylase and amylase-binding-protein A (AbpA) of Streptococcus gordonii with glucosyltransferase of S. gordonii and Streptococcus mutans.
Chaudhuri B, Rojek J, Vickerman MM, Tanzer JM, Scannapieco FA. Chaudhuri B, et al. BMC Microbiol. 2007 Jun 25;7:60. doi: 10.1186/1471-2180-7-60. BMC Microbiol. 2007. PMID: 17593303 Free PMC article. - Identification and analysis of a gene (abpA) encoding a major amylase-binding protein in Streptococcus gordonii.
Rogers JD, Haase EM, Brown AE, Douglas CWI, Gwynn JP, Scannapieco FA. Rogers JD, et al. Microbiology (Reading). 1998 May;144 ( Pt 5):1223-1233. doi: 10.1099/00221287-144-5-1223. Microbiology (Reading). 1998. PMID: 9611797 - Taking the starch out of oral biofilm formation: molecular basis and functional significance of salivary α-amylase binding to oral streptococci.
Nikitkova AE, Haase EM, Scannapieco FA. Nikitkova AE, et al. Appl Environ Microbiol. 2013 Jan;79(2):416-23. doi: 10.1128/AEM.02581-12. Epub 2012 Nov 9. Appl Environ Microbiol. 2013. PMID: 23144140 Free PMC article. Review. - In vitro models that support adhesion specificity in biofilms of oral bacteria.
Ellen RP, Lépine G, Nghiem PM. Ellen RP, et al. Adv Dent Res. 1997 Apr;11(1):33-42. doi: 10.1177/08959374970110011401. Adv Dent Res. 1997. PMID: 9524440 Review.
Cited by
- Uncovering Roles of Streptococcus gordonii SrtA-Processed Proteins in the Biofilm Lifestyle.
Nairn BL, Lee GT, Chumber AK, Steck PR, Mire MO, Lima BP, Herzberg MC. Nairn BL, et al. J Bacteriol. 2020 Dec 18;203(2):e00544-20. doi: 10.1128/JB.00544-20. Print 2020 Dec 18. J Bacteriol. 2020. PMID: 33106345 Free PMC article. - Interactions Between Streptococcus gordonii and Fusobacterium nucleatum Altered Bacterial Transcriptional Profiling and Attenuated the Immune Responses of Macrophages.
Liu T, Yang R, Zhou J, Lu X, Yuan Z, Wei X, Guo L. Liu T, et al. Front Cell Infect Microbiol. 2022 Jan 7;11:783323. doi: 10.3389/fcimb.2021.783323. eCollection 2021. Front Cell Infect Microbiol. 2022. PMID: 35071038 Free PMC article. - Effect of starch and amylase on the expression of amylase-binding protein A in Streptococcus gordonii.
Nikitkova AE, Haase EM, Scannapieco FA. Nikitkova AE, et al. Mol Oral Microbiol. 2012 Aug;27(4):284-94. doi: 10.1111/j.2041-1014.2012.00644.x. Epub 2012 Mar 14. Mol Oral Microbiol. 2012. PMID: 22759313 Free PMC article. - Draft genome sequences of 18 oral streptococcus strains that encode amylase-binding proteins.
Sabharwal A, Liao YC, Lin HH, Haase EM, Scannapieco FA. Sabharwal A, et al. Genome Announc. 2015 May 21;3(3):e00510-15. doi: 10.1128/genomeA.00510-15. Genome Announc. 2015. PMID: 25999552 Free PMC article. - The biogeography of infection revisited.
Azimi S, Lewin GR, Whiteley M. Azimi S, et al. Nat Rev Microbiol. 2022 Oct;20(10):579-592. doi: 10.1038/s41579-022-00683-3. Epub 2022 Feb 8. Nat Rev Microbiol. 2022. PMID: 35136217 Free PMC article. Review.
References
- Aguirre A, Levine M J, Cohen R E, Tabak L A. Immunochemical quantitation of α-amylase and secretory IgA in parotid saliva from people of various ages. Arch Oral Biol. 1987;32:297–301. - PubMed
- Behnke D. Plasmid transformation of Streptococcus sanguis (Challis) occurs by circular and linear molecules. Mol Gen Genet. 1981;182:490–497. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources