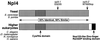The conserved npl4 protein complex mediates proteasome-dependent membrane-bound transcription factor activation - PubMed (original) (raw)
The conserved npl4 protein complex mediates proteasome-dependent membrane-bound transcription factor activation
A L Hitchcock et al. Mol Biol Cell. 2001 Oct.
Free PMC article
Abstract
Proteolytic activation of membrane-bound transcription factors has emerged as an important mechanism for the regulation of gene expression. Two membrane-bound transcription factors regulated in this manner are the Saccharomyces cerevisiae proteins Mga2p and Spt23p, which direct transcription of the Delta9-fatty acid desaturase gene OLE1. We now show that a membrane-associated complex containing the highly conserved Npl4p, Ufd1p, and Cdc48p proteins mediates the proteasome-regulated cleavage of Mga2p and Spt23p. Mutations in NPL4, UFD1, and CDC48 cause a block in Mga2p and Spt23p processing, with concomitant loss of OLE1 expression. Taken together, our data indicate that the Npl4 complex may serve to target the proteasome to the ubiquitinated endoplasmic reticulum membrane-bound proteins Mga2p and Spt23p. Given the recent finding that NPL4 is allelic to the ERAD gene HRD4, we further propose that this NPL4 function extends to all endoplasmic reticulum-membrane-associated targets of the proteasome.
Figures
Figure 1
The Npl4 protein is evolutionarily conserved. The sequence homology between S. cerevisiae_Npl4p protein (ScNpl4p) (accession number CAA85131), S. pombe predicted protein SPBC1711.10c (SpNpl4) (accession number CAB88240), C. elegans proteins F59E12.4 (CeNpl4[1]) (accession number AAB54254) and F59E12.5 (CeNpl4[2]) (accession number AAB54255), D. melanogaster predicted protein CG4673 (DmNpl4) (accession number AAF56480), R. norvegicus (rat) Npl4 (Meyer et al., 2000), and predicted H. sapiens (human) protein FLJ20657 (HsNpl4) (accession number NP 060391) is depicted schemati-cally. The_D. melanogaster Npl4 protein contains a large amino acid insertion omitted from this alignment. Ten absolutely conserved cysteine and histidine residues in the N terminus may constitute a novel Zn2+-finger domain. A C-terminal extension of higher eukaryotic Npl4 homologues encodes a predicted Nup153-like zinc-finger RanGDP-binding domain. The location of residues affected by mutations in the S. cerevisiae npl4-1 and npl4-2_mutants are indicated. The npl4-1 allele substitutes serine for the conserved glycine at position 323, and the_npl4-2 mutation results in the production of a truncated protein lacking the C-terminal 12 amino acids.
Figure 2
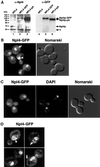
Npl4p-GFP localizes to ER/nuclear membranes. (A) Anti-Npl4 (left) and anti-GFP (right) Western blots of whole cell extracts derived from strains expressing endogenous Npl4p (lanes 1 and 4), a genomically tagged Npl4p-GFP (lanes 2 and 5), and a genomically tagged Npl4p-protein A(pA) (lanes 3 and 6). The anti-Npl4 blot shows the precise replacement of endogenous Npl4p with the two different C-terminally tagged proteins (compare lanes 2 and 3 to lane 1). The robust reactivity of anti-Npl4 with Npl4-pA (lane 3) is a result of antibody interactions with Npl4p epitopes as well as the IgG-binding pA tag. The rabbit anti-GFP blot recognizes both Npl4p-GFP and Npl4p-pA (lanes 5 and 6, respectively). The identity of the different proteins is indicated by arrows. The asterisk indicates an unidentified cross-reacting protein. (B) Npl4p-GFP localization as observed by fluorescence microscopy. The live cells were viewed at midlog phase. Localization is mostly nuclear (white arrowhead) and cytoplasmic, with an apparent concentration at perinuclear membranes. The white arrow indicates an example of Npl4p-GFP localization to peripheral membranes. A Nomarski image of the cells is shown to the right. (C) Npl4p-GFP colocalization with DAPI-stained nuclear DNA. Cells were fixed in formaldehyde, permeabilized with Triton X-100, and DAPI stained before fluorescence microscopy. The majority of Npl4p-GFP localizes to a region overlapping with and surrounding the nucleus as observed by DAPI staining. A Nomarski image of the cells is shown to the right. (D) Npl4p-GFP expression as viewed by fluorescence microscopy on a DeltaVision platform. Sequential 0.1-μm fluorescence images were taken through a diploid yeast strain expressing Npl4p-GFP from both_NPL4_ loci and subjected to five rounds of deconvolution. The images presented represent a central plane from deconvolved data. The white arrowhead indicates an example of Npl4p-GFP concentration at perinuclear membranes. White arrows indicate examples of Npl4p-GFP localization to peripheral membranes.
Figure 3
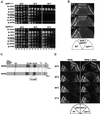
Biochemical characterization of Npl4p localization and interactions. (A) Npl4p is found in both soluble and membrane-bound fractions. Equal protein from crude whole cell extract (lane 1), purified microsomes (lane 2), and ER/nuclear membrane-cleared extract (lane 3) derived from a wild-type yeast strain was separated by SDS-PAGE and Western blotted with anti-Npl4 antibodies. The band corresponding to Npl4p is indicated, and asterisks denote unidentified cross-reacting proteins. (B) The microsomal fraction of Npl4p is tightly membrane associated. Purified microsomes from a wild-type yeast strain were washed with either buffer alone (lanes 1 and 2), 1 M KOAc (lanes 3 and 4), 2 M KOAc (lanes 5 and 6), 3 M urea (lanes 7 and 8), 0.1 M Na2CO3, pH 11 (lanes 9 and 10), or 1% Triton X-100 (lanes 11 and 12). The samples were then separated into pellet (P; membrane associated; odd lanes) or soluble (S; even lanes) fractions and subjected to SDS-PAGE and Western blotting with anti-Npl4 and anti-Cdc48 antibodies. The extraction profiles of the peripheral ER membrane protein Sec17p and the ER integral membrane protein Sec62p were determined by Western blotting with anti-Sec17 and anti-Sec62 antibodies as controls. (C) Npl4p-protein A (pA) copurifies with Cdc48p and Ufd1p. Yeast whole cell extracts derived from either a strain expressing untagged Npl4p (lane 1) or Npl4p-pA (lane 2) were incubated with IgG-Sepharose beads. After extensive washing, proteins bound to the beads were eluted with acid and visualized by SDS-PAGE and Coomassie blue staining. Mass spectral analysis of excised protein bands revealed the identity of the Npl4p-pA copurifying p100 and p43 proteins as Cdc48p and Ufd1p, respectively.
Figure 4
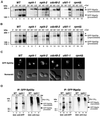
Yeast npl4 mutants are rescued by regulators of the UFA pathway and the proteasome. (A) Suppression of_npl4-1_ and npl4-2 temperature sensitivity by high-copy expression of OLE1, SPT23,MGA2, and RPN4. npl4-1 (PSY2340, top) and_npl4-2_ (PSY2341, bottom) strains transformed with either empty vector, or 2 μ vectors containing NPL4,OLE1, MGA2(aa1-914), MGA2,SPT23, or RPN4 were serially diluted onto YEPD media. The number of cells spotted in each dilution is indicated at the bottom. Plates were incubated at the indicated temperatures for 24–48 h. (B) Temperature sensitivity of a npl4-1 strain is extragenically suppressed by a transposon insertion allele of_SPT23_. Growth of wild-type (WT; FY23),npl4-1 (PSY2373), and npl4-1 SPT23:: Tn3:: LEU2 (PSY2377) strains on Leu−plates at the indicated temperatures. The FY23 and PSY2373 strains were transformed with an empty LEU2 vector to allow growth on Leu− dropout plates. (C) Schematic of the S. cerevisiae Mga2p and Spt23p proteins. These partially redundant factors are 38% identical and 54% similar. Sequence features include a putative basic nuclear localization signal (NLS), a putative leucine-rich nuclear export signal (NES), and two ankyrin repeats. The TIG domain is an Ig-like fold domain found in cell surface receptors and intracellular transcription factors including NF-κB. The C-terminal transmembrane domain (TM) is indicated. Blocks of strong amino acid conservation without assigned or predicted function are indicated by white boxes. The site of truncation in the isolated_MGA2_ high-copy suppressor clone (pPS2019) is indicated by an arrow in the Mga2p schematic. The site of transposon (Tn::LEU2) insertion in the_npl4-1_ extragenic suppressor allele of_SPT23_ (PSY2377) is indicated in the Spt23p schematic. (D) Temperature sensitivity of yeast npl4 mutants is partially rescued by supplementing growth media with UFAs. Wild-type (WT), ole1Δ, npl4-1, and_npl4-2_ strains were struck onto YEPD plates unsupplemented (left) or supplemented (right) with 0.5 mM UFAs (0.25 mM palmitoleic acid [16:1], 0.25 mM oleic acid [18:1], 1% Tergitol). Plates were incubated at the indicated temperatures for 48 h.
Figure 5
NPL4 function is required for_OLE1_ transcription. Northern analysis was performed on total RNA isolated from ole1Δ (lane 1), wild-type (WT; lanes 2–4), npl4-1 (lanes 5–7), npl4-2(lanes 8–10), cdc48-3 (lanes 11–13),ufd1-1 (lanes 14-16), and rpn4Δ cells (lanes 17–19) with OLE1_- and_ACT1_-specific DNA probes. Cells were grown in YEPD media at 25°C with or without supplemented UFAs. For wild-type cells, the temperature-sensitive-strains npl4-1,npl4-2, and cdc48-3, and non–temperature sensitive strains ufd1-1 and_rpn4Δ, samples were also collected after shifts to 37°C for 60 min. A normalized OLE1/_ACT1_ratio was calculated (see MATERIALS AND METHODS) for each sample to allow for quantitation of the relative changes in _OLE1_expression among samples.
Figure 6
NPL4 is required for efficient processing of ubiquitinated Mga2p and Spt23p fusion proteins. (A) Induction of GFP-Mga2p fusion protein in wild-type (WT; lanes 1–4),npl4-1 (lanes 5–7), npl4-2 (lanes 8–10), cdc48-2 (lanes 11–13), ufd1-1(lanes 14-16), and rpn4Δ (lanes 17–19) cells at 25°C. Cells were collected for analysis by anti-GFP Western blot at the indicated time points into galactose induction. Asterisks indicate higher molecular weight GFP-Mga2p proteins. (B) Induction of GFP-Spt23p fusion protein in wild-type (WT; lanes 1–4), npl4-1(lanes 5–7), npl4-2 (lanes 8–10),cdc48-2 (lanes 11–13), ufd1-1 (lanes 14-16), and rpn4Δ (lanes 17–19) cells at 25°C. Cells were collected for analysis by anti-GFP Western blot at the indicated time points into galactose induction. Asterisks indicate higher molecular weight GFP-Spt23p proteins. (C) Subcellular localization of GFP-Spt23p in wild-type (WT), npl4-1,npl4-2, cdc48-2, ufd1-1, and rpn4Δ cells after 120-min galactose inductions at 25°C. GFP-Spt23p signal is largely nucleoplasmic in wild-type cells, while remaining largely ER/nuclear envelope associated in the mutant strains. (D) GFP-Spt23p (left) and GFP-Mga2p (right) were immunoprecipitated (IP) from wild-type (WT) or _npl4-1_yeast whole cell extracts expressing Myc-tagged ubiquitin. Expression of GFP-Spt23p and GFP-Mga2p was either induced for 2 h with galactose (lanes 2, 4, 6, 8, 10, 12, 14, and 16), or repressed with the addition of glucose (lanes 1, 3, 5, 7, 9, 11, 13, and 15) before immunoprecipitation. Samples run on the same gel were analyzed by anti-GFP (lanes 1–4 and 9–12) or anti-Myc (lanes 5–8 and 13–16) Western blotting.
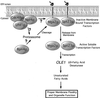
Figure 7
A model for Npl4 function in the proteasome-dependent processing of the ER membrane-bound transcription factors Mga2p and Spt23p. Details are presented in the text.
Similar articles
- HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins.
Bays NW, Wilhovsky SK, Goradia A, Hodgkiss-Harlow K, Hampton RY. Bays NW, et al. Mol Biol Cell. 2001 Dec;12(12):4114-28. doi: 10.1091/mbc.12.12.4114. Mol Biol Cell. 2001. PMID: 11739805 Free PMC article. - Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing.
Hoppe T, Matuschewski K, Rape M, Schlenker S, Ulrich HD, Jentsch S. Hoppe T, et al. Cell. 2000 Sep 1;102(5):577-86. doi: 10.1016/s0092-8674(00)00080-5. Cell. 2000. PMID: 11007476 - Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone.
Rape M, Hoppe T, Gorr I, Kalocay M, Richly H, Jentsch S. Rape M, et al. Cell. 2001 Nov 30;107(5):667-77. doi: 10.1016/s0092-8674(01)00595-5. Cell. 2001. PMID: 11733065 - Cdc48-Ufd1-Npl4: stuck in the middle with Ub.
Bays NW, Hampton RY. Bays NW, et al. Curr Biol. 2002 May 14;12(10):R366-71. doi: 10.1016/s0960-9822(02)00862-x. Curr Biol. 2002. PMID: 12015140 Review. - Yeast desaturases.
Martin CE, Oh CS, Kandasamy P, Chellapa R, Vemula M. Martin CE, et al. Biochem Soc Trans. 2002 Nov;30(Pt 6):1080-2. doi: 10.1042/bst0301080. Biochem Soc Trans. 2002. PMID: 12440977 Review.
Cited by
- Role of the ubiquitin-selective CDC48(UFD1/NPL4 )chaperone (segregase) in ERAD of OLE1 and other substrates.
Braun S, Matuschewski K, Rape M, Thoms S, Jentsch S. Braun S, et al. EMBO J. 2002 Feb 15;21(4):615-21. doi: 10.1093/emboj/21.4.615. EMBO J. 2002. PMID: 11847109 Free PMC article. - Ub-ProT reveals global length and composition of protein ubiquitylation in cells.
Tsuchiya H, Burana D, Ohtake F, Arai N, Kaiho A, Komada M, Tanaka K, Saeki Y. Tsuchiya H, et al. Nat Commun. 2018 Feb 6;9(1):524. doi: 10.1038/s41467-018-02869-x. Nat Commun. 2018. PMID: 29410401 Free PMC article. - Structure and function of the AAA+ ATPase p97/Cdc48p.
Xia D, Tang WK, Ye Y. Xia D, et al. Gene. 2016 May 25;583(1):64-77. doi: 10.1016/j.gene.2016.02.042. Epub 2016 Mar 3. Gene. 2016. PMID: 26945625 Free PMC article. Review. - Endoproteolytic cleavage of TUG protein regulates GLUT4 glucose transporter translocation.
Bogan JS, Rubin BR, Yu C, Löffler MG, Orme CM, Belman JP, McNally LJ, Hao M, Cresswell JA. Bogan JS, et al. J Biol Chem. 2012 Jul 6;287(28):23932-47. doi: 10.1074/jbc.M112.339457. Epub 2012 May 18. J Biol Chem. 2012. PMID: 22610098 Free PMC article. - Characterization of AtCDC48. Evidence for multiple membrane fusion mechanisms at the plane of cell division in plants.
Rancour DM, Dickey CE, Park S, Bednarek SY. Rancour DM, et al. Plant Physiol. 2002 Nov;130(3):1241-53. doi: 10.1104/pp.011742. Plant Physiol. 2002. PMID: 12427991 Free PMC article.
References
- Acharya U, Jacobs R, Peters JM, Watson N, Farquhar MG, Malhotra V. The formation of Golgi stacks from vesiculated Golgi membranes requires two distinct fusion events. Cell. 1995;82:895–904. - PubMed
- Adams A, Gottschling DE, Kaiser CA, Stearns T. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997.
- Agard DA, Hiraoka Y, Shaw P, Sedat JW. Fluorescence microscopy in three dimensions. Methods Cell Biol. 1989;30:353–377. - PubMed
- Agatep, R., Kirkpatrick, R.D., Parchaliuk, D.L., Woods, R.A., and Gietz, R.D. (1998). Transformation of Saccharomyces cerevisiae by the lithium acetate/single-stranded carrier DNA/polyethylene glycol (LiAc/ss-DNA/PEG) protocol: Technical Tips Online, http://tto.trends.com).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous