Control of specific gene expression by gibberellin and brassinosteroid - PubMed (original) (raw)
Comparative Study
Control of specific gene expression by gibberellin and brassinosteroid
T Bouquin et al. Plant Physiol. 2001 Oct.
Abstract
We identified a recessive, brassinolide-insensitive mutant caused by a deletion allele (bri1-201) of the brassinosteroid (BR) receptor BRI1. The bri1-201 mutant displayed altered expression levels of genes differentially regulated by gibberellin (GA). RNA-blot analysis revealed that BR and GA antagonistically regulate the accumulation of mRNAs of the GA-responsive GASA1 gene, as well as the GA-repressible GA5 gene. Expression studies with cycloheximide indicated that the antagonistic effects of GA and BR on GA5 require de novo protein synthesis. Reporter transgene analyses and RNA-blot analysis showed that BR and GA modulate GA5 expression, at least in part, at the transcriptional level, and that the signals are independent and subtractive.
Figures
Figure 1
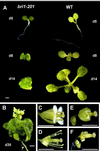
Phenotype of the bri1-201 mutant under long-day conditions (16 h light/8 h dark). A, Comparative development of bri1-201 and WT. B, bri1-201 35 d after germination. C through F, WT (C and D) and bri1-201 flowers (E and F). In D and F, sepals and petals were removed to show reproductive organs. Bars represent 1 mm.
Figure 2
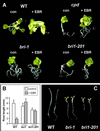
bri1-201 exhibits insensitivity to EBR and constitutive skotomorphogenesis. A, Two-week-old WT and bri1-201 were transferred to plates without (con) or with (+EBR) 0.5 μ
m
EBR and grown for 5 d. B, Root length of WT, bri1-1 and bri1-201 seedlings grown 5 d without (control) or with (+EBR) 0.2 μ
m
EBR. Each value represents the mean of 50 independent measurements. C, Skotomorphogenesis of WT and mutant seedlings. Seedlings were germinated and grown for 5 d in darkness on Murashige and Skoog medium.
Figure 3
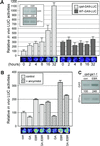
RNA-blot analysis of GA5, GA4, GASA1, GAI, and RGA mRNA accumulation in bri1-201 and WT upon GA3 treatment. Poly(A+) RNA (1 μg lane−1) from 16-d-old plants grown on Murashige and Skoog medium without or with 50 μ
m
GA3 added for the last 48 h. Ribonucleic 32P-CTP antisense probes were synthesized using T7 RNA polymerase from partial cDNA 3′ sequences cloned in the pGEM-Teasy vector. GA5, GA4, and GASA1 hybridizations were performed on the same filter, whereas GAI and RGA hybridizations were performed on independent filters. Radioactive signals were quantified on all membranes and standardized (WT con = 100) by comparison to signals obtained after subsequent blotting with the EF1-α probe.
Figure 4
GASA1 and GA5 mRNA accumulation is antagonistically controlled by GA and BR. Two-week-old seedlings grown on Murashige and Skoog were transferred to 50 mL liquid one-half-strength Murashige and Skoog in flasks for 1 d prior to treatment. RNA-blot analyses were performed and normalized with EF1-α as described in Figure 3. A, WT, bri1-201 and cpd seedlings were treated with 1 μ
m
EBR for 48 h. B, cpd seedlings were treated with GA or BR in presence (CHX) or not of translational inhibitors. CHX (50 μ
m
) and chloramphenicol (50 μ
m
) were added to the medium 2 h before GA or BR treatments (50 μ
m
GA3 or 1 μ
m
EBR for 16 h).
Figure 5
Effects of BR and GA on GA5-LUC expression. A, LUC imaging of the _GA5_-LUC reporter in 16-d-old WT-_GA5_-LUC and _cpd_-_GA5_-LUC seedlings treated with 1 μ
m
EBR for various times. Insert in A, GA5 transcript accumulation monitored by RNA-blot analysis from 16-d-old cpd seedlings treated with 1 μ
m
EBR for 2 h. Hybridization conditions and normalization with EF1-α were performed as described in Figure 3. B, One-week-old cpd-GA5-LUC seedlings grown on Murashige and Skoog plates were transferred to liquid one-half-strength Murashige and Skoog in presence or absence of ancymidol (1 mg L−1) and grown for 7 more d. BR (0.1 μ
m
EBR) and GA (50 μ
m
GA3) treatments were performed for 16 h by adding the hormones to the medium. C, GA5 transcript accumulation in 16-d-old cpd-ga1-1 double mutant seedlings treated with 1 μ
m
EBR for 16 h. Hybridization conditions and normalization with EF1-α were performed as described in Figure 3. In A and B, bioluminescent LUC images (displayed in pseudocolors) acquired by CCD camera are shown below the quantification (average gray values) of LUC images.
Similar articles
- Arabidopsis RAV1 is down-regulated by brassinosteroid and may act as a negative regulator during plant development.
Hu YX, Wang YX, Liu XF, Li JY. Hu YX, et al. Cell Res. 2004 Feb;14(1):8-15. doi: 10.1038/sj.cr.7290197. Cell Res. 2004. PMID: 15040885 - A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development.
Clouse SD, Langford M, McMorris TC. Clouse SD, et al. Plant Physiol. 1996 Jul;111(3):671-8. doi: 10.1104/pp.111.3.671. Plant Physiol. 1996. PMID: 8754677 Free PMC article. - GCR1 can act independently of heterotrimeric G-protein in response to brassinosteroids and gibberellins in Arabidopsis seed germination.
Chen JG, Pandey S, Huang J, Alonso JM, Ecker JR, Assmann SM, Jones AM. Chen JG, et al. Plant Physiol. 2004 Jun;135(2):907-15. doi: 10.1104/pp.104.038992. Epub 2004 Jun 4. Plant Physiol. 2004. PMID: 15181210 Free PMC article. - The brassinosteroid signal transduction pathway.
Wang ZY, Wang Q, Chong K, Wang F, Wang L, Bai M, Jia C. Wang ZY, et al. Cell Res. 2006 May;16(5):427-34. doi: 10.1038/sj.cr.7310054. Cell Res. 2006. PMID: 16699538 Free PMC article. Review. - Ligand perception, activation, and early signaling of plant steroid receptor brassinosteroid insensitive 1.
Jiang J, Zhang C, Wang X. Jiang J, et al. J Integr Plant Biol. 2013 Dec;55(12):1198-211. doi: 10.1111/jipb.12081. Epub 2013 Sep 9. J Integr Plant Biol. 2013. PMID: 23718739 Review.
Cited by
- The microRNA159-regulated GAMYB-like genes inhibit growth and promote programmed cell death in Arabidopsis.
Alonso-Peral MM, Li J, Li Y, Allen RS, Schnippenkoetter W, Ohms S, White RG, Millar AA. Alonso-Peral MM, et al. Plant Physiol. 2010 Oct;154(2):757-71. doi: 10.1104/pp.110.160630. Epub 2010 Aug 10. Plant Physiol. 2010. PMID: 20699403 Free PMC article. - Brassinosteroid-regulated bHLH transcription factor CESTA induces the gibberellin 2-oxidase GA2ox7.
Albertos P, Wlk T, Griffiths J, Pimenta Lange MJ, Unterholzner SJ, Rozhon W, Lange T, Jones AM, Poppenberger B. Albertos P, et al. Plant Physiol. 2022 Mar 28;188(4):2012-2025. doi: 10.1093/plphys/kiac008. Plant Physiol. 2022. PMID: 35148416 Free PMC article. - Comparative Transcriptome Analysis Points to the Biological Processes of Hybrid Incompatibility between Brassica napus and B. oleracea.
Yue F, Zheng F, Li Q, Mei J, Shu C, Qian W. Yue F, et al. Plants (Basel). 2023 Jul 12;12(14):2622. doi: 10.3390/plants12142622. Plants (Basel). 2023. PMID: 37514237 Free PMC article. - HORMONOMETER: a tool for discerning transcript signatures of hormone action in the Arabidopsis transcriptome.
Volodarsky D, Leviatan N, Otcheretianski A, Fluhr R. Volodarsky D, et al. Plant Physiol. 2009 Aug;150(4):1796-805. doi: 10.1104/pp.109.138289. Epub 2009 Jun 17. Plant Physiol. 2009. PMID: 19535475 Free PMC article. - A Quantitative Proteomic Analysis of Brassinosteroid-induced Protein Phosphorylation in Rice (Oryza sativa L.).
Hou Y, Qiu J, Wang Y, Li Z, Zhao J, Tong X, Lin H, Zhang J. Hou Y, et al. Front Plant Sci. 2017 Apr 7;8:514. doi: 10.3389/fpls.2017.00514. eCollection 2017. Front Plant Sci. 2017. PMID: 28439285 Free PMC article.
References
- Axelos M, Bardet C, Liboz T, Le Van Thai A, Curie C, Lescure B. The gene family encoding the Arabidopsis thaliana translation elongation factor EF-1 alpha: molecular cloning, characterization and expression. Mol Gen Genet. 1989;219:106–112. - PubMed
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases