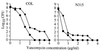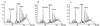Role of penicillin-binding protein 4 in expression of vancomycin resistance among clinical isolates of oxacillin-resistant Staphylococcus aureus - PubMed (original) (raw)
Role of penicillin-binding protein 4 in expression of vancomycin resistance among clinical isolates of oxacillin-resistant Staphylococcus aureus
J E Finan et al. Antimicrob Agents Chemother. 2001 Nov.
Abstract
It has been reported that penicillin-binding protein 4 (PBP4) activity decreases when a vancomycin-susceptible Staphylococcus aureus isolate is passaged in vitro to vancomycin resistance. We analyzed the PBP profiles of four vancomycin intermediately susceptible S. aureus (VISA) clinical isolates and found that PBP4 was undetectable in three isolates (HIP 5827, HIP 5836, and HIP 6297) and markedly reduced in a fourth (Mu50). PBP4 was readily visible in five vancomycin-susceptible, oxacillin-resistant S. aureus (ORSA) isolates. The nucleotide sequences of the pbp4 structural gene and flanking sequences did not different between the VISA and vancomycin-susceptible isolates. Overproduction of PBP4 on a high-copy-number plasmid in the VISA isolates produced a two- to threefold decrease in vancomycin MICs. Inactivation of pbp4 by allelic replacement mutagenesis in three vancomycin-susceptible ORSA strains (COL, RN450M, and N315) led to a decrease in vancomycin susceptibility, an increase in highly vancomycin-resistant subpopulations, and decreased cell wall cross-linking by high-performance liquid chromatography analysis. Complementation of the COL mutant with plasmid-encoded pbp4 restored the vancomycin MIC and increased cell wall cross-linking. These data suggest that alterations in PBP4 expression are at least partially responsible for the VISA phenotype.
Figures
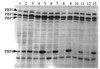
FIG. 1
PBP profiles in the presence and absence of vancomycin. Lanes: 1, RN450M without vancomycin; 2, HIP5827 with vancomycin; 3, HIP5827 without vancomycin; 4, HIP5836 with vancomycin; 5, HIP5836 without vancomycin; 6, Mu50 with vancomycin; 7, Mu50 without vancomycin; 8, Mu3 with vancomycin; 9, Mu3 without vancomycin.
FIG. 2
PBP profiles of control strains RN4220 (lane 1), RN4220/pJF3 (lane 2), and RN450M (lane 3) and VISA isolates Mu3 (lane 4), Mu3/pJF3 (lane 5), Mu50 (lane 6), Mu50/pJF3 (lane 7), HIP6297 (lane 8), HIP6297/pJF3 (lane 9), HIP 5836 (lane 10), HIP5836/pJF3 (lane 11), HIP5827 (lane 12), and HIP5827/pJF3 (lane 13).
FIG. 3
EOP curves for HIP5836 and HIP5827. Shown are numbers of_S. aureus_ bacteria (in log10 CFU per milliliter) remaining on plates containing various concentrations of vancomycin. Each parental VISA isolate is represented as a solid square. Each parental VISA isolate containing the cloning vector alone (pSK265) is represented as a solid circle. Each VISA isolate expressing_pbp4_ on high-copy plasmid pJF3 is represented as a solid diamond.
FIG. 4
EOP curves for pbp4 knockouts of COL and N315. Shown on the y axis are the numbers of_S. aureus_ bacteria (in log10 CFU per milliliter) remaining on the plates containing various concentrations of vancomycin. Parent strains N315 and COL are represented by solid squares, and their isogenic pbp4 knockouts are represented by solid circles.
FIG. 5
Analysis of peptidoglycan from S. aureus COL (A), COL with pbp4 inactivated following allelic mutagenesis (B), and COL with pbp4_inactivated complemented with pJF3 (C) by reverse-phase HPLC. COL with_pbp4 inactivated demonstrates a lower degree of muropeptide cross-linking, as evidenced by the lower number of oligomeric muropeptides at the end of the chromatogram. Complementation of the inactivated pbp4 gene with pJF3 leads to a higher degree of cross-linking, as evidenced by the increase in trimers and oligomers in panel C. mAU, milliabsorption units.
FIG. 6
Vancomycin gradient plate of COL and COLΔ_pbp4_ before and after overnight exposure to vancomycin at 1 μg/ml. Lanes: 1, COL with no exposure; 2, COL after overnight exposure to vancomycin; 3, COLΔ_pbp4_ with no exposure; 4, COLΔ_pbp4_ after overnight exposure to vancomycin.
Similar articles
- Transcriptional induction of the penicillin-binding protein 2 gene in Staphylococcus aureus by cell wall-active antibiotics oxacillin and vancomycin.
Boyle-Vavra S, Yin S, Challapalli M, Daum RS. Boyle-Vavra S, et al. Antimicrob Agents Chemother. 2003 Mar;47(3):1028-36. doi: 10.1128/AAC.47.3.1028-1036.2003. Antimicrob Agents Chemother. 2003. PMID: 12604538 Free PMC article. - Inactivated pbp4 in highly glycopeptide-resistant laboratory mutants of Staphylococcus aureus.
Sieradzki K, Pinho MG, Tomasz A. Sieradzki K, et al. J Biol Chem. 1999 Jul 2;274(27):18942-6. doi: 10.1074/jbc.274.27.18942. J Biol Chem. 1999. PMID: 10383392 - Detection of intrinsic oxacillin resistance in non-multiresistant, oxacillin-resistant Staphylococcus aureus (NORSA).
Gosbell IB, Neville SA, Mercer JL, Fernandes LA, Fernandes CJ. Gosbell IB, et al. J Antimicrob Chemother. 2003 Feb;51(2):468-70. doi: 10.1093/jac/dkg064. J Antimicrob Chemother. 2003. PMID: 12562728 No abstract available. - [The mechanism of vancomycin-resistant in Staphylococcus aureus].
Kuroda M, Hiramatsu K. Kuroda M, et al. Tanpakushitsu Kakusan Koso. 2000 Jun;45(8):1329-38. Tanpakushitsu Kakusan Koso. 2000. PMID: 10846471 Review. Japanese. No abstract available. - Molecular mechanisms of methicillin resistance in Staphylococcus aureus.
Domínguez MA, Liñares J, Martín R. Domínguez MA, et al. Microbiologia. 1997 Sep;13(3):301-8. Microbiologia. 1997. PMID: 9353748 Review.
Cited by
- tcaA inactivation increases glycopeptide resistance in Staphylococcus aureus.
Maki H, McCallum N, Bischoff M, Wada A, Berger-Bächi B. Maki H, et al. Antimicrob Agents Chemother. 2004 Jun;48(6):1953-9. doi: 10.1128/AAC.48.6.1953-1959.2004. Antimicrob Agents Chemother. 2004. PMID: 15155184 Free PMC article. - Divergent Effects of Peptidoglycan Carboxypeptidase DacA on Intrinsic β-Lactam and Vancomycin Resistance.
Park SH, Choi U, Ryu SH, Lee HB, Lee JW, Lee CR. Park SH, et al. Microbiol Spectr. 2022 Aug 31;10(4):e0173422. doi: 10.1128/spectrum.01734-22. Epub 2022 Jun 27. Microbiol Spectr. 2022. PMID: 35758683 Free PMC article. - Selection of heterogeneous vancomycin-intermediate Staphylococcus aureus by imipenem.
Katayama Y, Murakami-Kuroda H, Cui L, Hiramatsu K. Katayama Y, et al. Antimicrob Agents Chemother. 2009 Aug;53(8):3190-6. doi: 10.1128/AAC.00834-08. Epub 2009 May 18. Antimicrob Agents Chemother. 2009. PMID: 19451283 Free PMC article. - A mecA-negative strain of methicillin-resistant Staphylococcus aureus with high-level β-lactam resistance contains mutations in three genes.
Banerjee R, Gretes M, Harlem C, Basuino L, Chambers HF. Banerjee R, et al. Antimicrob Agents Chemother. 2010 Nov;54(11):4900-2. doi: 10.1128/AAC.00594-10. Epub 2010 Aug 30. Antimicrob Agents Chemother. 2010. PMID: 20805396 Free PMC article. - Staphylococcus aureus PBP4 is essential for beta-lactam resistance in community-acquired methicillin-resistant strains.
Memmi G, Filipe SR, Pinho MG, Fu Z, Cheung A. Memmi G, et al. Antimicrob Agents Chemother. 2008 Nov;52(11):3955-66. doi: 10.1128/AAC.00049-08. Epub 2008 Aug 25. Antimicrob Agents Chemother. 2008. PMID: 18725435 Free PMC article.
References
- Climo M W, Markowitz S, Williams D, Stuart G, Archer G. Comparison of the in vitro and in vivo efficacy of FKO37, vancomycin, imipenem, and nafcillin against staphylococcal species. J Antimicrob Chemother. 1997;40:59–66. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous