p300 acts as a transcriptional coactivator for mammalian Notch-1 - PubMed (original) (raw)
p300 acts as a transcriptional coactivator for mammalian Notch-1
F Oswald et al. Mol Cell Biol. 2001 Nov.
Abstract
Notch-1 belongs to a family of transmembrane receptor proteins that direct the decisions as to various cell fates. After ligand binding, a proteolytic cleavage step occurs and the intracellular part of Notch-1, Notch-1-IC, translocates into the nucleus, where it targets the DNA binding protein RBP-J kappa/CBF1. RBP-J kappa mediates repression through recruitment of a histone deacetylase-containing complex. The Notch-1-IC/RBP-J kappa complex overcomes repression and activates the transcription of Notch target genes. We have identified a novel domain in Notch-1-IC, the EP domain, which is indispensable for full transcriptional activation. This transactivation domain is localized adjacent to the ankyrin repeats of Notch-1-IC. In cotransfection experiments, Notch-1-IC-mediated transcriptional activation was inhibited by E1A12S and p53, two proteins, which interfere with the function of the common coactivator p300. Protein-protein interaction assays demonstrated the association of Notch-1-IC and the CH3 region of p300. In addition, the interaction of mammalian Notch-1-IC with p300 was destabilized after deletion of the EP domain of Notch-1-IC. Based on physical interaction with Notch-1-IC and coactivator functions of p300, we propose a model for Notch-1-mediated gene regulation via p300.
Figures
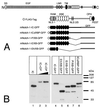
FIG. 1
Schematic representation of reporter and mNotch-1-IC-specific expression constructs. (A) The luciferase construct pGa981/6 carries 12 RBP-Jκ binding sites upstream of a minimal β-globin promoter. Abbreviations: H3, _Hin_dIII, N, _Nsi_I, X, _Xho_I. (B) The mammalian Notch-1 receptor and the derived deletion proteins used for expression. All constructs carry an N-terminal FLAG tag (open circle). The first and last amino acids compared to the full-length protein are indicated in parentheses. The −ΔEP mutants represent in-frame deletions of aa 2098 to 2112. The asterisk designates a 1758 WFP/LAA 1760 mutation in the mRAM23 domain. Abbreviations: SS, signal sequence, EGF, epidermal growth factor-like repeats, LNR, Lin/Notch repeats, TM, transmembrane domain, RAM, RAM23 domain, ANK, ankyrin repeats, NLS, nuclear localization signal.
FIG. 2
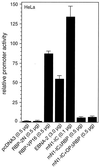
Notch-1-IC-mediated transcription depends on the primary RBP-Jκ binding site within the RAM domain. Portions (2 μg) of reporter construct pGa981/6 were cotransfected into HeLa cells together with the indicated amounts of expression plasmids. Luciferase activity was determined from 100-μg portions of total-cell extracts, and the basal promoter activity of the reporter construct was set to unity. Mean values and standard deviations from at least four independent experiments are shown.
FIG. 3
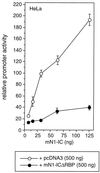
Dominant negative effect of mNotch-1-ICΔRBP. Portions (2 μg) of reporter construct pGa981/6 were cotransfected into HeLa cells together with increasing amounts of a Notch-1-IC expression plasmid and the indicated amount of pcDNA3 (open circles) or pcDNA3mN1-ICΔRBP (solid circles). Luciferase activity was determined from 100-μg portions of total-cell extracts, and the basal promoter activity of the reporter construct was set to unity. Mean values and standard deviations from three independent experiments are shown.
FIG. 4
Transcriptional activity of mNotch-1 deletion mutants. Portions (2 μg) of reporter construct pGa981/6 were cotransfected into HeLa cells with 150 or 200 ng of plasmid expressing the indicated mNotch-1 proteins. Luciferase activity was determined from 100-μg portions of total-cell extracts, and the basal promoter activity of the reporter construct was set to unity. Mean values and standard deviations from at least five independent experiments are shown.
FIG. 5
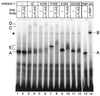
Detection of RBP/Notch-1 DNA binding complexes. In cell extracts from HEK-293 cells, an RBP-Jκ-specific DNA binding complex can be detected (complex A, lanes 1 and 2). In cell extracts from HEK-293 cells transfected with expression plasmids for the indicated Notch-1 proteins, the RBP-Jκ-specific DNA binding activity was decreased and more slowly migrating complexes appeared (lanes 3 to 12). Transcriptionally active Notch-1 proteins form a slowly migrating complex (complex C, lanes 3, 5, and 7). Treatment with an anti-FLAG antibody (α-FLAG) recognized complex C and supershifted to a novel complex D (lanes 4, 6, and 8). Transcriptionally inactive Notch-1 proteins form a faster-migrating complex (complex E, lanes 9 and 11). In cell extracts from HEK-293 cells transfected with the Notch-1 mutant mNotch-1-P/XB, both complexes (C and D, lanes 7) were formed. Complex E interfered with the anti-FLAG antibody (lanes 8, 10, and 12). Cell-free synthesized RBP-2N was used as a control for RBP DNA binding. The RBP-specific DNA binding activity (A) (lane 13) was recognized by an RBP-specific antibody (α-RBP) to form a supershifted complex B (lane 14). The asterisk designates a nonspecific complex (lanes 3 to 12). The 32P-labeled oligonucleotide SL233 was used as a probe.
FIG. 6
(A to C) Transcriptional activity of Notch-1–GFP fusion proteins. (A) Schematic representation of Notch-1 deletion mutants fused to GFP. For details, see Fig. 1B. (B) Cell extracts were prepared 24 h after transfection of 5-μg portions of plasmid expressing Notch-1 proteins and Notch-1-specific GFP fusion proteins. Expression was assayed by Western blotting using an anti-FLAG antibody. (C) Stimulation of the reporter construct pGa981/6 by Notch-1–GFP fusion proteins depends on the EP domain. Portions (2 μg) of reporter construct pGa981/6 were cotransfected into HeLa cells with 150, 200, or 250 ng of plasmid expressing the indicated mNotch-1-GFP fusion proteins. Luciferase activity was determined from 100-μg portions of total-cell extracts, and the basal promoter activity of the reporter construct was set to unity. Mean values and standard deviations of four independent experiments are shown. (D) Subcellular localization of Notch-1-GFP fusion proteins in vivo. HEK-293 cells were transfected with plasmids expressing GFP (CMV-GFP) (a) or the Notch-1-IC-derived GFP fusion proteins mNotch-1-IC-GFP (b), mNotch-1-ΔRBP-GFP (c), mNotch-1-P/XB-GFP (d), mNotch-1-E/XB-GFP (e), or mNotch-1-Eh/XB-GFP (f). At 24 h after transfection, the living cells were assayed for GFP expression by fluorescence microscopy. Magnification, ×630.
FIG. 6
(A to C) Transcriptional activity of Notch-1–GFP fusion proteins. (A) Schematic representation of Notch-1 deletion mutants fused to GFP. For details, see Fig. 1B. (B) Cell extracts were prepared 24 h after transfection of 5-μg portions of plasmid expressing Notch-1 proteins and Notch-1-specific GFP fusion proteins. Expression was assayed by Western blotting using an anti-FLAG antibody. (C) Stimulation of the reporter construct pGa981/6 by Notch-1–GFP fusion proteins depends on the EP domain. Portions (2 μg) of reporter construct pGa981/6 were cotransfected into HeLa cells with 150, 200, or 250 ng of plasmid expressing the indicated mNotch-1-GFP fusion proteins. Luciferase activity was determined from 100-μg portions of total-cell extracts, and the basal promoter activity of the reporter construct was set to unity. Mean values and standard deviations of four independent experiments are shown. (D) Subcellular localization of Notch-1-GFP fusion proteins in vivo. HEK-293 cells were transfected with plasmids expressing GFP (CMV-GFP) (a) or the Notch-1-IC-derived GFP fusion proteins mNotch-1-IC-GFP (b), mNotch-1-ΔRBP-GFP (c), mNotch-1-P/XB-GFP (d), mNotch-1-E/XB-GFP (e), or mNotch-1-Eh/XB-GFP (f). At 24 h after transfection, the living cells were assayed for GFP expression by fluorescence microscopy. Magnification, ×630.
FIG. 6
(A to C) Transcriptional activity of Notch-1–GFP fusion proteins. (A) Schematic representation of Notch-1 deletion mutants fused to GFP. For details, see Fig. 1B. (B) Cell extracts were prepared 24 h after transfection of 5-μg portions of plasmid expressing Notch-1 proteins and Notch-1-specific GFP fusion proteins. Expression was assayed by Western blotting using an anti-FLAG antibody. (C) Stimulation of the reporter construct pGa981/6 by Notch-1–GFP fusion proteins depends on the EP domain. Portions (2 μg) of reporter construct pGa981/6 were cotransfected into HeLa cells with 150, 200, or 250 ng of plasmid expressing the indicated mNotch-1-GFP fusion proteins. Luciferase activity was determined from 100-μg portions of total-cell extracts, and the basal promoter activity of the reporter construct was set to unity. Mean values and standard deviations of four independent experiments are shown. (D) Subcellular localization of Notch-1-GFP fusion proteins in vivo. HEK-293 cells were transfected with plasmids expressing GFP (CMV-GFP) (a) or the Notch-1-IC-derived GFP fusion proteins mNotch-1-IC-GFP (b), mNotch-1-ΔRBP-GFP (c), mNotch-1-P/XB-GFP (d), mNotch-1-E/XB-GFP (e), or mNotch-1-Eh/XB-GFP (f). At 24 h after transfection, the living cells were assayed for GFP expression by fluorescence microscopy. Magnification, ×630.
FIG. 7
Repression of Notch-1-mediated transactivation by E1A12S and p53. (A and B) Portions (2 μg) of reporter construct pGa981/6 were cotransfected into HeLa cells with 100 ng of plasmid expressing mNotch-1-IC+OP (A) or mNotch-1-IC (B), together with increasing amounts (0.2, 0.4, and 0.6 μg) of plasmids expressing the indicated E1A proteins. (C) Portions (2 μg) of reporter construct pGa981/6 were cotransfected with 100-ng portions of plasmid expressing mNotch-1-IC+OP together with increasing amounts (0.2, 0.4, and 0.6 μg) of plasmids expressing the indicated human and murine p53 proteins. Luciferase activity was determined from 100-μg portions of total-cell extracts, and the basal promoter activity of the reporter construct was set to unity. Mean values and standard deviations from four independent experiments are shown.
FIG. 8
Notch-1-IC interacts with the CH3 region of p300. (A) Schematic representation of p300-derived protein fragments fused to GST. After bacterial expression, the GST fusion proteins were used in pull-down experiments. The first and last amino acids compared to the full-length protein are indicated in parentheses. (B) Cell-free synthesized E1A protein interacts with various p300 fragments. (C) Cell-free synthesized Notch-1-IC protein interacts with p300 fragment B. (D) Cell-free synthesized p300 interacts with GSTmNotch-1-IC. C-terminally truncated p300 (aa 1 to 1302) fails to interact with GST-Notch-1-IC. GST proteins were immobilized with Sepharose beads and incubated with in vitro-translated and radiolabeled proteins. After extensive washing steps, the reaction mixtures were boiled and the proteins were separated by SDS-polyacrylamide gel electrophoresis. The positions of molecular size markers are shown.
FIG. 9
(A) Notch-1-IC/p300 interaction correlates with transcriptional activity. (A) GST p300 fragment B was immobilized with Sepharose beads and incubated with in vitro-translated Notch-1-IC deletion mutants (TNT-N1-) or E1A protein as a control. After extensive washing steps, the reaction mixtures were boiled and proteins were separated by SDS-polyacrylamide gel electrophoresis. Relative transcriptional activity of the Notch-1 proteins in cotransfection experiments is shown (Fig. 4). ++, strong transcriptional activation; +, reduced activation; −, loss of activation. (B) Relative binding of in vitro-translated Notch-1-IC deletion mutants to GST-p300 fragment B obtained from densitometric analysis. Exposed films from six experiments were scanned and analyzed by the NIH Image software.
FIG. 9
(A) Notch-1-IC/p300 interaction correlates with transcriptional activity. (A) GST p300 fragment B was immobilized with Sepharose beads and incubated with in vitro-translated Notch-1-IC deletion mutants (TNT-N1-) or E1A protein as a control. After extensive washing steps, the reaction mixtures were boiled and proteins were separated by SDS-polyacrylamide gel electrophoresis. Relative transcriptional activity of the Notch-1 proteins in cotransfection experiments is shown (Fig. 4). ++, strong transcriptional activation; +, reduced activation; −, loss of activation. (B) Relative binding of in vitro-translated Notch-1-IC deletion mutants to GST-p300 fragment B obtained from densitometric analysis. Exposed films from six experiments were scanned and analyzed by the NIH Image software.
FIG. 10
Notch-1/p300 interaction in vivo. Expression of endogenous p300, mNotch-1-IC+OP (lane 1), and mNotch-1-IC+OP-ΔEP (lane 2) in transiently transfected HEK-293 cells is shown. The amount of p300 protein in cell lysates (lane 3) decreased after incubation with the anti-p300 antibody (lane 4) and accumulated in the precipitate (lane 6). Increasing amounts of blocking peptide added to the reaction mixture (lanes 8 and 9) prevented immunoprecipitation of p300 as well as coimmunoprecipitation of mNotch-1-IC+OP (lane 7). Deletion of the EP domain destabilizes Notch-1/p300 interaction (lanes 10 to 15). Increasing amounts of LiCl in the washing buffer did not interfere with p300 binding to mNotch-1-IC+OP but destabilized p300 binding to mNotch-1-IC+OP-ΔEP (compare lanes 10 to 12 with lanes 13 to 15). Extracts were incubated with agarose beads alone (lane 5) or an agarose-conjugated anti-p300 antibody (lanes 6 to 15). The mixture was divided into three aliquots, and the beads were washed three times with CHAPS lysis buffer containing 150 mM (lanes 10 and 13), 500 mM (lanes 11 and 14), or 1000 mM LiCl (lanes 12 and 15). After a further washing step with CHAPS lysis buffer, the proteins were analyzed by Western blotting.
FIG. 11
Mutation analysis of the EP domain within mNotch-1-IC. (A) Schematic representation of the mNotch-1-IC specific scanning mutants used in cotransfection experiments. (B) Transcriptional activity of EP domain mutants. Portions (2 μg) of reporter construct pGa981/6 were cotransfected into HeLa cells with 150-ng portions of plasmid expressing the indicated mNotch-1-IC proteins. Luciferase activity was determined from 100-μg portions of total-cell extracts, and the basal promoter activity of the reporter construct was set to unity. (C) Cell extracts from HEK-293 cells were prepared 24 h after transfection of 5 μg of plasmid expressing Notch-1-IC–EP domain mutant proteins. Expression was assayed by Western blotting using an anti-FLAG antibody. Mean values and standard deviations from four independent experiments are shown. (D) The EP domain is highly conserved within Notch proteins. Protein sequences of the indicated Notch proteins were aligned with the CLUSTAL W program. The first and last amino acids are numbered relative to the full-length proteins. The EP domain is specified. Abbreviations and accession numbers: N-dros, Notch Drosophila (P07207); N1-xen, Notch-1 Xenopus laevis (P21783); TAN-1, Notch-1 human (P46531); N1-mm, Notch-1 mouse (Q01705); N2-mm, Notch-2 mouse (BAA22094); N3-mm, Notch-3 mouse (Q61982).
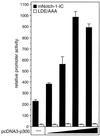
FIG. 12
Additive effect of p300 in Notch-1-mediated transactivation. Portions (2 μg) of reporter construct pGa981/6 were cotransfected into HeLa cells with 100 ng of plasmids expressing mNotch-1-IC (black bars) or the mNotch-1 specific EP domain mutant LDE/AAA (white bars), together with increasing amounts (50, 100, 200, and 500 ng) of pcDNA3-p300. Luciferase activity was determined from 100-μg portions of total-cell extracts, and the basal promoter activity of the reporter construct was set to unity. Mean values and standard deviations of three independent experiments are shown.
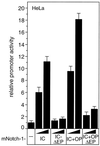
FIG. 13
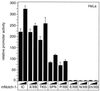
Transcriptional activation of the murine HES-1 promoter by Notch-1 depends on the EP domain. Portions (2 μg) of the HES-1 specific reporter construct HES-1-LUC were cotransfected into HeLa cells with 150 or 200 ng of plasmid expressing the indicated mNotch-1 proteins. Luciferase activity was determined from 100-μg portions of total-cell extracts, and the basal promoter activity of the reporter construct was set to unity. Mean values and standard deviations from at least three independent experiments are shown.
Similar articles
- p300 and PCAF act cooperatively to mediate transcriptional activation from chromatin templates by notch intracellular domains in vitro.
Wallberg AE, Pedersen K, Lendahl U, Roeder RG. Wallberg AE, et al. Mol Cell Biol. 2002 Nov;22(22):7812-9. doi: 10.1128/MCB.22.22.7812-7819.2002. Mol Cell Biol. 2002. PMID: 12391150 Free PMC article. - SHARP is a novel component of the Notch/RBP-Jkappa signalling pathway.
Oswald F, Kostezka U, Astrahantseff K, Bourteele S, Dillinger K, Zechner U, Ludwig L, Wilda M, Hameister H, Knöchel W, Liptay S, Schmid RM. Oswald F, et al. EMBO J. 2002 Oct 15;21(20):5417-26. doi: 10.1093/emboj/cdf549. EMBO J. 2002. PMID: 12374742 Free PMC article. - The notch 3 intracellular domain represses notch 1-mediated activation through Hairy/Enhancer of split (HES) promoters.
Beatus P, Lundkvist J, Oberg C, Lendahl U. Beatus P, et al. Development. 1999 Sep;126(17):3925-35. doi: 10.1242/dev.126.17.3925. Development. 1999. PMID: 10433920 - Linking Notch signaling, chromatin remodeling, and T-cell leukemogenesis.
Bresnick EH, Chu J, Christensen HM, Lin B, Norton J. Bresnick EH, et al. J Cell Biochem Suppl. 2000;Suppl 35:46-53. doi: 10.1002/1097-4644(2000)79:35+<46::aid-jcb1125>3.0.co;2-5. J Cell Biochem Suppl. 2000. PMID: 11389531 Review. - The shortest path from the surface to the nucleus: RBP-J kappa/Su(H) transcription factor.
Honjo T. Honjo T. Genes Cells. 1996 Jan;1(1):1-9. doi: 10.1046/j.1365-2443.1996.10010.x. Genes Cells. 1996. PMID: 9078362 Review.
Cited by
- Cutaneous papillomavirus E6 oncoproteins associate with MAML1 to repress transactivation and NOTCH signaling.
Brimer N, Lyons C, Wallberg AE, Vande Pol SB. Brimer N, et al. Oncogene. 2012 Oct 25;31(43):4639-46. doi: 10.1038/onc.2011.589. Epub 2012 Jan 16. Oncogene. 2012. PMID: 22249263 Free PMC article. - Chromatin signatures at Notch-regulated enhancers reveal large-scale changes in H3K56ac upon activation.
Skalska L, Stojnic R, Li J, Fischer B, Cerda-Moya G, Sakai H, Tajbakhsh S, Russell S, Adryan B, Bray SJ. Skalska L, et al. EMBO J. 2015 Jul 14;34(14):1889-904. doi: 10.15252/embj.201489923. Epub 2015 Jun 11. EMBO J. 2015. PMID: 26069324 Free PMC article. - Rhamnetin and cirsiliol induce radiosensitization and inhibition of epithelial-mesenchymal transition (EMT) by miR-34a-mediated suppression of Notch-1 expression in non-small cell lung cancer cell lines.
Kang J, Kim E, Kim W, Seong KM, Youn H, Kim JW, Kim J, Youn B. Kang J, et al. J Biol Chem. 2013 Sep 20;288(38):27343-27357. doi: 10.1074/jbc.M113.490482. Epub 2013 Jul 31. J Biol Chem. 2013. PMID: 23902763 Free PMC article. - MAML1 acts cooperatively with EGR1 to activate EGR1-regulated promoters: implications for nephrogenesis and the development of renal cancer.
Hansson ML, Behmer S, Ceder R, Mohammadi S, Preta G, Grafström RC, Fadeel B, Wallberg AE. Hansson ML, et al. PLoS One. 2012;7(9):e46001. doi: 10.1371/journal.pone.0046001. Epub 2012 Sep 27. PLoS One. 2012. PMID: 23029358 Free PMC article. - Notch activation stimulates transient and selective binding of Su(H)/CSL to target enhancers.
Krejcí A, Bray S. Krejcí A, et al. Genes Dev. 2007 Jun 1;21(11):1322-7. doi: 10.1101/gad.424607. Genes Dev. 2007. PMID: 17545467 Free PMC article.
References
- Arias J, Alberts A S, Brindle P, Claret F X, Smeal T, Karin M, Feramisco J, Montminy M. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature. 1994;370:226–229. - PubMed
- Artavanis-Tsakonas S, Rand M D, Lake R J. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. - PubMed
- Aster J C, Robertson E S, Hasserjian R P, Turner J R, Kieff E, Sklar J. Oncogenic forms of NOTCH1 lacking either the primary binding site for RBP-Jκ or nuclear localization sequences retain the ability to associate with RBP-Jκ and activate transcription. J Biol Chem. 1997;272:11336–11343. - PubMed
- Bailey A M, Posakony J W. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 1995;9:2609–2622. - PubMed
- Beatus P, Lundkvist J, Oberg C, Lendahl U. The notch 3 intracellular domain represses notch 1-mediated activation through Hairy/Enhancer of split (HES) promoters. Development. 1999;126:3925–3935. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous