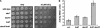A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation - PubMed (original) (raw)
A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation
R Swanson et al. Genes Dev. 2001.
Abstract
Substrate discrimination in the ubiquitin-proteasome system is believed to be dictated by specific combinations of ubiquitin-protein ligases (E3s) and ubiquitin-conjugating enzymes (E2s). Here we identify Doa10/Ssm4 as a yeast E3 that is embedded in the endoplasmic reticulum (ER)/nuclear envelope yet can target the soluble transcription factor Matalpha2. Doa10 contains an unusual RING finger, which has ubiquitin-ligase activity in vitro and is essential in vivo for degradation of alpha2 via its Deg1 degradation signal. Doa10 functions with two E2s, Ubc6 and Ubc7, to ubiquitinate Deg1-bearing substrates, and it is also required for the degradation of at least one ER membrane protein. Interestingly, different short-lived ER proteins show distinct requirements for Doa10 and another ER-localized E3, Hrd1. Nevertheless, the two E3s overlap in function: A doa10Delta hrd1Delta mutant is far more sensitive to cadmium relative to either single mutant and displays strong constitutive induction of the unfolded protein response; this suggests a role for both E3s in eliminating aberrant ER proteins. The likely human ortholog of DOA10 is in the cri-du-chat syndrome critical region on chromosome 5p, suggesting that defective ubiquitin ligation might contribute to this common genetic disorder.
Figures
Figure 1
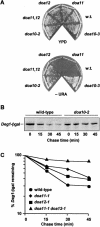
Characterization of new doa mutants. (A) Wild-type and doa mutant strains were grown on rich medium (YPD) and minimal plates lacking uracil (−URA). (B) Pulse-chase analysis of _Deg1_–βgal in wild-type and doa10-2 strains. (C) Degradation kinetics of Deg1-βgal in wild-type, doa11-1, doa12-1, and doa11-1 doa12-1 strains. Proteins were precipitated with antibodies to βgal for B and C.
Figure 2
Doa10 structural features. (A) Alignment of the closely related yeast Doa10 and human TEB4 RING fingers with examples of a RING-HC finger (human c-Cbl) and a RING-H2 finger (yeast Hrd1). The metal-coordinating His and Cys residues are highlighted in gray, as is the Trp residue commonly found in RING finger ubiquitin ligases. An 11-residue segment after residue 359 of Hrd1 was removed for clarity. (†) Residues in the c-Cbl RING that contact the E2 in the c-Cbl–UbcH7 structure (Zheng et al. 2000). (B) The Doa10 WW domain compared to two other yeast non-type-I WW motifs. The consensus sequence is from Kasanov et al. (2001). (δ) hydrophilic; (Θ) aromatic. (C) Similar membrane topology predicted from hydropathy plots of S. cerevisiae Doa10 and its likely orthologs in S. pombe (SPBC14F5.07) and humans (TEB4; GenBank KIAA0597). The predicted human protein lacks several internal, poorly conserved segments shared by the two yeast proteins. (D) The TEB4–Doa10 (TD) domain. The same proteins as in C are compared.
Figure 3
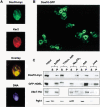
Doa10 is an integral membrane protein of the ER/nuclear envelope. (A) MHY1657 cells expressing myc9-tagged Doa10 were stained with an antibody to the myc epitope, an antibody to Kar2, and the Hoechst 33258 dye. (B) GFP fluorescence visualized by confocal microscopy in MHY1658 cells expressing a fusion between Doa10 and GFP. (C) Subcellular fractionation of MHY1690 cells carrying pHA-UBC7. Cell lysates were divided into microsomal pellet and supernatant fractions, which were examined by immunoblotting. Lysates were treated with buffer alone or buffer containing 1% Triton X-100 and 0.5 M NaCl (Triton, salt), 0.1 M Na2CO3 at pH 11.5, 0.5 M NaCl (salt), or 2.5 M urea. (P) pellet; (S) supernatant.
Figure 4
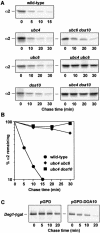
Doa10 shows high substrate specificity in vivo. (A) Degradation kinetics of the N-end rule substrate Leu–βgal in wild-type (MHY501) and _doa10_Δ strains (MHY1631) carrying the plasmid p415GPD-Ub-L-lacZ. Radiolabeled proteins were precipitated with an antibody to βgal. (B) Degradation kinetics of CPY* in wild-type and mutant strains. Radiolabeled proteins were precipitated with an antibody to CPY. (C) Degradation of Ubc6. At time zero, cycloheximide was added, and disappearance of Ubc6 was followed by anti-Ubc6 immunoblotting.
Figure 5
Matα2 degradation in mutant cells. (A) Pulse-chase analysis of α2 in wild-type cells and congenic deletion mutants. Proteins were precipitated with an antibody to α2. (B) Quantitation of the pulse-chase data in A for MHY501, MHY503, and MHY1648. (C) _Deg1_–βgal degradation in wild-type cells (MHY501) carrying the plasmid YEp13–Deg1–lacZ and either full-length DOA10 under the control of a strong promoter on a 2-μm plasmid (pGPD–DOA10) or the empty vector (pGPD). Proteins were precipitated with an antibody to βgal.
Figure 6
Function of the Doa10 RING finger in vivo and in vitro. (A) Anti-ubiquitin immunoblot analysis of immunoprecipitated _Deg1_–βgal from cells carrying YEplac195–_Deg1_–lacZ or empty vector. Strains also contained a plasmid for overexpression of ubiquitin (+Ub) or an empty vector. Proteins were immunoprecipitated with antibodies to βgal. (Lower panel) The same blot reprobed with antibodies against βgal. (B) Degradation of _Deg1_–βgal in wild-type and doa10–C39S cells. Proteins were precipitated with an antibody to βgal. (C) In vitro ubiquitin ligase activity of the Doa10 RING domain. (Lanes 1,2) Complete reactions, including S protein substrate. In the reaction with S peptide, 1.5 μg of competitor peptide was added (lane 2). Ubiquitin–S protein conjugates (Ubn-S) are indicated. Omitted components are indicated above lanes 5–9, and no S protein was added in the zinc chelation experiment shown in lanes 3 and 4. Ubiquitinated proteins were detected by anti-ubiquitin immunoblotting. Open arrowhead indicates position of unmodified S protein.
Figure 7
Overlap in function of the ER-localized Doa10 and Hrd1 ubiquitin ligases. (A) The _doa10_Δ _hrd1_Δ double mutant is hypersensitive to cadmium. Congenic cells of the indicated genotypes were spotted in 10-fold dilutions on minimal medium containing CdCl2 or on YPD and incubated for 4 d and 2 d at 30°C, respectively. (B) Induction of the unfolded protein response in a _hrd1_Δ _doa10_Δ mutant. Congenic cells carried the plasmid pSZ1, which contains a UPRE–lacZ reporter (Friedlander et al. 2000).
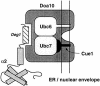
Figure 8
Model for Doa10 function. See text for details.
Similar articles
- A Conserved C-terminal Element in the Yeast Doa10 and Human MARCH6 Ubiquitin Ligases Required for Selective Substrate Degradation.
Zattas D, Berk JM, Kreft SG, Hochstrasser M. Zattas D, et al. J Biol Chem. 2016 Jun 3;291(23):12105-18. doi: 10.1074/jbc.M116.726877. Epub 2016 Apr 11. J Biol Chem. 2016. PMID: 27068744 Free PMC article. - Spatially regulated ubiquitin ligation by an ER/nuclear membrane ligase.
Deng M, Hochstrasser M. Deng M, et al. Nature. 2006 Oct 19;443(7113):827-31. doi: 10.1038/nature05170. Nature. 2006. PMID: 17051211 - N-terminal acetylation of the yeast Derlin Der1 is essential for Hrd1 ubiquitin-ligase activity toward luminal ER substrates.
Zattas D, Adle DJ, Rubenstein EM, Hochstrasser M. Zattas D, et al. Mol Biol Cell. 2013 Apr;24(7):890-900. doi: 10.1091/mbc.E12-11-0838. Epub 2013 Jan 30. Mol Biol Cell. 2013. PMID: 23363603 Free PMC article. - Natural substrates of the proteasome and their recognition by the ubiquitin system.
Ulrich HD. Ulrich HD. Curr Top Microbiol Immunol. 2002;268:137-74. doi: 10.1007/978-3-642-59414-4_6. Curr Top Microbiol Immunol. 2002. PMID: 12083004 Review. - The DOA pathway: studies on the functions and mechanisms of ubiquitin-dependent protein degradation in the yeast Saccharomyces cerevisiae.
Hochstrasser M, Papa FR, Chen P, Swaminathan S, Johnson P, Stillman L, Amerik AY, Li SJ. Hochstrasser M, et al. Cold Spring Harb Symp Quant Biol. 1995;60:503-13. doi: 10.1101/sqb.1995.060.01.054. Cold Spring Harb Symp Quant Biol. 1995. PMID: 8824423 Review. No abstract available.
Cited by
- The SUD1 gene encodes a putative E3 ubiquitin ligase and is a positive regulator of 3-hydroxy-3-methylglutaryl coenzyme a reductase activity in Arabidopsis.
Doblas VG, Amorim-Silva V, Posé D, Rosado A, Esteban A, Arró M, Azevedo H, Bombarely A, Borsani O, Valpuesta V, Ferrer A, Tavares RM, Botella MA. Doblas VG, et al. Plant Cell. 2013 Feb;25(2):728-43. doi: 10.1105/tpc.112.108696. Epub 2013 Feb 12. Plant Cell. 2013. PMID: 23404890 Free PMC article. - Quality control of cytoplasmic proteins inside the nucleus.
Borgert L, Mishra S, den Brave F. Borgert L, et al. Comput Struct Biotechnol J. 2022 Aug 23;20:4618-4625. doi: 10.1016/j.csbj.2022.08.033. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36090811 Free PMC article. Review. - Growth-based determination and biochemical confirmation of genetic requirements for protein degradation in Saccharomyces cerevisiae.
Watts SG, Crowder JJ, Coffey SZ, Rubenstein EM. Watts SG, et al. J Vis Exp. 2015 Feb 16;(96):e52428. doi: 10.3791/52428. J Vis Exp. 2015. PMID: 25742191 Free PMC article. - Variation in ubiquitin system genes creates substrate-specific effects on proteasomal protein degradation.
Collins MA, Mekonnen G, Albert FW. Collins MA, et al. Elife. 2022 Oct 11;11:e79570. doi: 10.7554/eLife.79570. Elife. 2022. PMID: 36218234 Free PMC article. - Degradation Signals for Ubiquitin-Proteasome Dependent Cytosolic Protein Quality Control (CytoQC) in Yeast.
Maurer MJ, Spear ED, Yu AT, Lee EJ, Shahzad S, Michaelis S. Maurer MJ, et al. G3 (Bethesda). 2016 Jul 7;6(7):1853-66. doi: 10.1534/g3.116.027953. G3 (Bethesda). 2016. PMID: 27172186 Free PMC article.
References
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York, NY: Wiley; 1989.
- Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY. Hrd1p/ Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat Cell Biol. 2001;3:24–29. - PubMed
- Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin–proteasome system by protein aggregation. Science. 2001;292:1552–1555. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials