The hairless gene mutated in congenital hair loss disorders encodes a novel nuclear receptor corepressor - PubMed (original) (raw)
The hairless gene mutated in congenital hair loss disorders encodes a novel nuclear receptor corepressor
G B Potter et al. Genes Dev. 2001.
Abstract
The mammalian hairless (hr) gene plays a critical role in the maintenance of hair growth. Although the hr gene has been identified, the biochemical function of its encoded protein (Hr) has remained obscure. Here, we show that Hr functions as a transcriptional corepressor for thyroid hormone receptors (TRs). We find that two independent regions of Hr mediate TR binding and that interaction requires a cluster of hydrophobic residues similar to the binding motifs proposed for nuclear receptor corepressors (N-CoR and SMRT). Similarly, we show that Hr binds to the same region of TR as known corepressors. We show that Hr interacts with histone deacetylases (HDACs) and is localized to matrix-associated deacetylase (MAD) bodies, indicating that the mechanism of Hr-mediated repression is likely through associated HDAC activity. Thus, Hr is a component of the corepressor machinery, and despite its lack of sequence identity with previously described corepressors, its mode of action is remarkably conserved. On the basis of its thyroid hormone-inducible and tissue- and developmental-specific expression, Hr likely defines a new class of nuclear receptor corepressors that serve a more specialized role than ubiquitous corepressors. The discovery that Hr is a corepressor provides a molecular basis for specific hair loss syndromes in both humans and mice.
Figures
Figure 1
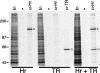
Hr interacts with TR in vivo. COS cells transfected with expression vectors for TRα and Hr both individually and together (Hr + TR) were metabolically labeled with [35S]methionine and protein extracts were prepared and used for immunoprecipitation analysis. Hr-specific antiserum (α-Hr) immunoprecipitates Hr (closed arrowhead) and, in the presence of TR, coprecipitates TR (open arrowhead). Immunoprecipitation with TR-specific antiserum (α-TR) shows position of TR (open arrowhead). (In) Two percent of extract used for immunoprecipitation. Sizes of molecular mass markers (kD) are indicated.
Figure 2
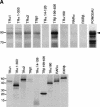
Mapping of the interaction domains of Hr with TR. (A) Deletion derivatives of Hr tested in the Far Western assay for interaction with TR. Bacterially expressed fusion proteins of various Hr deletion derivatives were separated by SDS-PAGE, transferred to nitrocellulose, and incubated with 35S-labeled TRβ1. (Left) Autoradiograph of Far Western filter; (right) Ponceau red staining to show loading of bacterial protein extracts and sizes of fusion proteins. (B) Summary of yeast two-hybrid (2H) and Far Western (FW) assay results. Yeast two-hybrid results were scored by growth on medium lacking histidine; (+) growth equal to 568–1207, (+/−) positive but weaker growth, (−) no detectable growth. All experiments were repeated at least twice with the same results. (C) Hr interacts with TR in the absence of ligand. Derivatives of Hr encoding TR interaction domains tested in the Far Western assay of duplicate blots incubated with 35S-labeled TRβ1 in the absence (−T3) or presence (+T3) of 10−6 M L-T3. Derivative 864–981 is included as a negative control. (Left and middle) Autoradiographs of Far Western filters; (right) Ponceau red stain. (D) Coimmunoprecipitation of individually expressed TR-IDs. COS cells were cotransfected with expression vectors for TRα and epitope-tagged Hr deletion derivatives that included the indicated amino acids. Protein extracts were used for immunoprecipitation analysis with TR-specific antiserum (α-TR) or mouse IgG (negative control). Hr derivatives were detected by Western analysis with Myc-specific antiserum. (Right) Western analysis of extracts with TR-specific antiserum to show expression of TR. Hr fragments expressed in COS cells alone did not precipitate with TR-specific antiserum (data not shown). (In) One percent of extract used for immunoprecipitation except for 750–1085 (5%). Sizes of molecular mass markers (in kD) are indicated.
Figure 3
Hydrophobic amino acids in TR-ID1 mediate Hr–TR interaction. (A) Region of homology between Hr and TRIP8. Identical residues are indicated by dashes. (B) Far Western analysis of Hr mutants. (Top) Residues in TR-ID1 changed to alanine by site-directed mutagenesis (within deletion derivative 568–864) indicated by black boxes. Regions predicted to form alpha-helical secondary structure are underlined. (Left) Autoradiograph of Far Western filter; (right) Ponceau red staining to show equivalent loading of bacterial protein extracts. (Arrowhead) Position of mutated Hr fusion proteins. (C) Summary of yeast two-hybrid (2H) and Far Western (FW) assays for interaction of mutated Hr derivatives. Yeast two-hybrid results were scored by growth on medium lacking histidine. (+) Growth equal to 568–864; (+/−) positive but weaker growth; (−) no detectable growth. For Far Western analysis: (+) signal equal to 568–864; (+/−) significantly weaker signal than 568–864; (−) no detectable signal. All experiments were repeated at least twice with the same results.
Figure 4
Analysis of Hr carboxy-terminal interaction domain (TR-ID2). (A) Far Western analysis of deletion derivatives from Hr amino acids 980–1084. (Top left) autoradiograph of Far Western filter; (top right) Ponceau red staining to show equivalent loading of bacterial protein extracts. (Open arrowheads) Positions of Hr fusion proteins. (Bottom) Schematic representation of deletion derivatives from Hr amino acids 980–1084. The minimal region in both interacting dervatives is between amino acids 1024 and 1040. (B) Far Western analysis of point mutants in TR-ID2. Site-directed mutatgenesis was used to change the indicated residues to alanine (within deletion derivative 980–1084). Underlined residues are predicted to form alpha-helical secondary structure. (Left) Autoradiograph of Far Western filter; (right) Ponceau red staining to show equivalent loading of bacterial protein extracts. (Arrowhead) Position of mutated Hr fusion proteins, mutant M11 migrates slightly slower than other mutants; (++) interaction equivalent to 980–1084; (+) interaction weaker than 980–1084; (+/−) weak but detectable interaction; (−) no detectable interaction. (C) Summary of site-directed mutagenesis results for Hr TR-IDs. Boxed residues showed no interaction when changed to alanine, underlined residues showed partial interaction. Below is shown a comparison of Hr consensus with consensus sequences defined for receptor binding by N-CoR and SMRT. Consensus 1 is from Nagy et al. (1999), Hu and Lazar (1999), and Webb et al. (2000); consensus 2 is from Perissi et al. (1999). Bold residues indicate Hr consensus extended to match consensus 2.
Figure 4
Analysis of Hr carboxy-terminal interaction domain (TR-ID2). (A) Far Western analysis of deletion derivatives from Hr amino acids 980–1084. (Top left) autoradiograph of Far Western filter; (top right) Ponceau red staining to show equivalent loading of bacterial protein extracts. (Open arrowheads) Positions of Hr fusion proteins. (Bottom) Schematic representation of deletion derivatives from Hr amino acids 980–1084. The minimal region in both interacting dervatives is between amino acids 1024 and 1040. (B) Far Western analysis of point mutants in TR-ID2. Site-directed mutatgenesis was used to change the indicated residues to alanine (within deletion derivative 980–1084). Underlined residues are predicted to form alpha-helical secondary structure. (Left) Autoradiograph of Far Western filter; (right) Ponceau red staining to show equivalent loading of bacterial protein extracts. (Arrowhead) Position of mutated Hr fusion proteins, mutant M11 migrates slightly slower than other mutants; (++) interaction equivalent to 980–1084; (+) interaction weaker than 980–1084; (+/−) weak but detectable interaction; (−) no detectable interaction. (C) Summary of site-directed mutagenesis results for Hr TR-IDs. Boxed residues showed no interaction when changed to alanine, underlined residues showed partial interaction. Below is shown a comparison of Hr consensus with consensus sequences defined for receptor binding by N-CoR and SMRT. Consensus 1 is from Nagy et al. (1999), Hu and Lazar (1999), and Webb et al. (2000); consensus 2 is from Perissi et al. (1999). Bold residues indicate Hr consensus extended to match consensus 2.
Figure 4
Analysis of Hr carboxy-terminal interaction domain (TR-ID2). (A) Far Western analysis of deletion derivatives from Hr amino acids 980–1084. (Top left) autoradiograph of Far Western filter; (top right) Ponceau red staining to show equivalent loading of bacterial protein extracts. (Open arrowheads) Positions of Hr fusion proteins. (Bottom) Schematic representation of deletion derivatives from Hr amino acids 980–1084. The minimal region in both interacting dervatives is between amino acids 1024 and 1040. (B) Far Western analysis of point mutants in TR-ID2. Site-directed mutatgenesis was used to change the indicated residues to alanine (within deletion derivative 980–1084). Underlined residues are predicted to form alpha-helical secondary structure. (Left) Autoradiograph of Far Western filter; (right) Ponceau red staining to show equivalent loading of bacterial protein extracts. (Arrowhead) Position of mutated Hr fusion proteins, mutant M11 migrates slightly slower than other mutants; (++) interaction equivalent to 980–1084; (+) interaction weaker than 980–1084; (+/−) weak but detectable interaction; (−) no detectable interaction. (C) Summary of site-directed mutagenesis results for Hr TR-IDs. Boxed residues showed no interaction when changed to alanine, underlined residues showed partial interaction. Below is shown a comparison of Hr consensus with consensus sequences defined for receptor binding by N-CoR and SMRT. Consensus 1 is from Nagy et al. (1999), Hu and Lazar (1999), and Webb et al. (2000); consensus 2 is from Perissi et al. (1999). Bold residues indicate Hr consensus extended to match consensus 2.
Figure 5
Mapping of the TR–Hr interaction domain. Deletion derivatives of TR were tested in the yeast two-hybrid and Far Western assays for interaction with Hr. (A) TR derivatives tested in Far Western assay. Protein extract from bacteria expressing GST-568–1207 was separated by SDS-PAGE, transferred to nitrocellulose, and individual lanes incubated with 35S-labeled TR derivatives (see schematic in B) as indicated. Panels are autoradiographs of Far Western filters. (Far right) Ponceau red staining to show loading of extract. (Arrowhead) Position of GST-568–1207. (Bottom) SDS–polyacrylamide gel showing 35S-labeled in vitro translated proteins used as probes. (B) Summary of yeast two-hybrid (2H) and Far West-ern (FW) assays for interaction of TR derivatives. Yeast two-hybrid results were scored by growth on medium lacking histidine. (+) Interaction; (−) no interaction; (ND) not done. Shaded region of TRα2 indicates a divergent carboxyl terminus; TRα160 has a point mutation changing a proline residue at position 160 to arginine. (DNA) DNA-binding domain; (T3) hormone-binding domain; (AF-2) coactivator-interaction domain in helix 12. All experiments were repeated at least twice with the same results.
Figure 5
Mapping of the TR–Hr interaction domain. Deletion derivatives of TR were tested in the yeast two-hybrid and Far Western assays for interaction with Hr. (A) TR derivatives tested in Far Western assay. Protein extract from bacteria expressing GST-568–1207 was separated by SDS-PAGE, transferred to nitrocellulose, and individual lanes incubated with 35S-labeled TR derivatives (see schematic in B) as indicated. Panels are autoradiographs of Far Western filters. (Far right) Ponceau red staining to show loading of extract. (Arrowhead) Position of GST-568–1207. (Bottom) SDS–polyacrylamide gel showing 35S-labeled in vitro translated proteins used as probes. (B) Summary of yeast two-hybrid (2H) and Far West-ern (FW) assays for interaction of TR derivatives. Yeast two-hybrid results were scored by growth on medium lacking histidine. (+) Interaction; (−) no interaction; (ND) not done. Shaded region of TRα2 indicates a divergent carboxyl terminus; TRα160 has a point mutation changing a proline residue at position 160 to arginine. (DNA) DNA-binding domain; (T3) hormone-binding domain; (AF-2) coactivator-interaction domain in helix 12. All experiments were repeated at least twice with the same results.
Figure 6
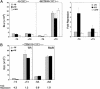
Hr mediates transcriptional repression by unliganded TR. (A) TH-responsive (MLV TRE-tk-luc) and control (tk-luc) reporter genes were cotransfected into GH1 cells together with an expression vector for Hr (+Hr) or expression vector alone (−Hr). Luciferase activity was measured in the absence (−T3) and presence (+T3) of 10–7 M L-T3. ΔHr is a Hr derivative that lacks both TR-IDs and consists of amino acids 1–568.Relative luciferase activity (RLU) is luciferase activity normalized to β-gal activity from CMX β-gal. Shown is the mean of two experiments done in duplicate. (Right) Fold repression is luciferase activity in the absence of Hr divided by activity in the presence of Hr. Shown are the mean values from three experiments done in duplicate. (B) Transcriptional repression by Hr is specific for TR. A TH-responsive reporter gene which can be activated by both TR and retinoic acid receptor (RAR) (TREpal-tk-luc) was cotransfected into GC cells together with an expression vector for TR (left) or RAR (right). The effect of Hr on transcription by these receptors was assayed by cotransfection of an expression vector for Hr (+Hr) or expression vector only (−Hr). Luciferase activity was measured in the absence and presence of 10–7 M L-T3 (T3) or 10–6 M all-trans retinoic acid (RA). Relative luciferase activity (RLU) is luciferase activity normalized to β-gal activity from CMX β-gal. Experiments were done three times with similar results; shown is a representative experiment. Fold repression is luciferase activity in the absence of Hr divided by activity in the presence of Hr and is the mean of three experiments done in duplicate.
Figure 7
Hr has multiple independent repression domains. (A) Fusion proteins of Hr with the GAL4 DNA-binding domain (DBD) were used in a cotransfection assay with a GAL4-responsive promoter (GALp3-tk luc). Values were calculated by normalization of luciferase activity to β-gal activity from CMX β-gal; fold repression is activity of GAL4 DBD divided by the activity of the indicated deletion derivative. Results are the mean of four independent experiments done in duplicate. (B) Schematic representation of Hr functional domains defined for TR interaction (TR-ID) and transcriptional repression (RD). Repression domains are shaded, ZF indicates region proposed to form a zinc finger. (TR-ID1) Amino acids 816–830; (TR-ID2) amino acids 827–838; (RD1) amino acids 236–450; (RD2) amino acids 568–864; (RD3) amino acids 864–981.
Figure 8
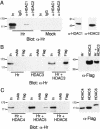
Hr associates with HDACs. (A) Hr associates with endogenous HDAC1. (Left) Protein extracts prepared from COS cells transfected with an expression vector for Hr (Hr) or vector only (Mock) were used for immunoprecipitation with HDAC1- and HDAC2-specific antisera. Hr was detected by Western analysis with Hr-specific antisera. (In) 10% of extract used for immunoprecipitation; (IgG) nonspecific rabbit IgG (negative control). (Right) Western analysis detecting HDAC1 and HDAC2 in extract used for immunoprecipitation (10% of input). (B) Hr coimmunoprecipitates with HDAC3. (Left) Protein extracts prepared from COS cells transfected with expression vectors for Hr, FLAG-tagged HDAC3, or both (Hr + HDAC3). FLAG-specific antiserum or a nonspecific monoclonal antiserum (mAb) were used for immunoprecipitation. Hr was detected by Western analysis with Hr-specific antisera. (In) Ten percent of extract used for immunoprecipitation. (Right) Western analysis of extract used for immunoprecipitation with FLAG-specific antisera to detect HDAC3. (C) Hr associates with HDAC5. (Left) COS cells were doubly transfected with expression vectors for Hr and FLAG-tagged HDAC4–HDAC6. FLAG-specific antiserum or a nonspecific monoclonal antibody (mAb) were used for immunoprecipitation. Hr was detected by Western analysis with Hr-specific antiserum. (Right) Western analysis of extracts used for immunoprecipation with FLAG-specific antiserum to detect HDACs. Protein loaded is 10% of amount used for immunoprecipitation. Sizes of molecular mass markers (kD) are indicated.
Figure 9
Hr localizes to MAD bodies. (A) Hr localizes to subnuclear structures. COS cells were transfected with an expression vector for Hr, plated on coverslips and used for indirect immunofluorescence with Hr-specific antiserum. (Left) DAPI staining to show nucleus; (center) staining with Hr-specific antiserum detected with Cy3-conjugated secondary antiserum; (right) differential interference contrast (DIC) image of cell. Bar, 20 μm. (B) Hr and HDAC5 colocalize to the same subnuclear bodies. COS cells were transfected with expression vectors for Hr (Hr), FLAG-tagged HDAC5 (HDAC5), or both (Hr + HDAC5) and detected by indirect immunofluorescence with Hr- or FLAG-specific antiserum. α-Hr and α-FLAG were detected with Cy3- and FITC-conjugated secondary antisera, respectively. Merge, overlayed images from Cy3 and FITC; yellow color indicates regions of overlap. (C) Hr colocalizes with SMRT in MAD bodies. COS cells were transfected with the indicated expression vectors and used for indirect immunofluorescence. (Top) Hr and SMRT detected by use of myc- and SMRT-specific antisera, respectively. α-myc and α-SMRT were detected with Cy3- and FITC-conjugated antisera, respectively. Merge, overlayed images from Cy3 and FITC; yellow indicates regions of overlap. (Middle) SMRT and HDAC5 detected with SMRT- or FLAG-specific antiserum. α-SMRT and α-FLAG were detected with Cy3- and FITC-conjugated secondary antisera, respectively. Merge, overlayed images from Cy3 and FITC; yellow indicates regions of overlap. (Bottom) Hr and RNA splicing factor SC-35 detected with Hr- and SC-35-specific antisera. α-Hr and α-SC-35 were detected with Cy3- and FITC-conjugated secondary antisera, respectively. Merge, overlayed images from Cy3 and FITC; yellow indicates regions of overlap.
Similar articles
- The specificity of interactions between nuclear hormone receptors and corepressors is mediated by distinct amino acid sequences within the interacting domains.
Cohen RN, Brzostek S, Kim B, Chorev M, Wondisford FE, Hollenberg AN. Cohen RN, et al. Mol Endocrinol. 2001 Jul;15(7):1049-61. doi: 10.1210/mend.15.7.0669. Mol Endocrinol. 2001. PMID: 11435607 - The corepressor N-CoR and its variants RIP13a and RIP13Delta1 directly interact with the basal transcription factors TFIIB, TAFII32 and TAFII70.
Muscat GE, Burke LJ, Downes M. Muscat GE, et al. Nucleic Acids Res. 1998 Jun 15;26(12):2899-907. doi: 10.1093/nar/26.12.2899. Nucleic Acids Res. 1998. PMID: 9611234 Free PMC article. - Characterization of receptor interaction and transcriptional repression by the corepressor SMRT.
Li H, Leo C, Schroen DJ, Chen JD. Li H, et al. Mol Endocrinol. 1997 Dec;11(13):2025-37. doi: 10.1210/mend.11.13.0028. Mol Endocrinol. 1997. PMID: 9415406 - N-CoR-HDAC corepressor complexes: roles in transcriptional regulation by nuclear hormone receptors.
Jones PL, Shi YB. Jones PL, et al. Curr Top Microbiol Immunol. 2003;274:237-68. doi: 10.1007/978-3-642-55747-7_9. Curr Top Microbiol Immunol. 2003. PMID: 12596910 Review. - Corepressor requirement and thyroid hormone receptor function during Xenopus development.
Sachs LM. Sachs LM. Vitam Horm. 2004;68:209-30. doi: 10.1016/S0083-6729(04)68007-1. Vitam Horm. 2004. PMID: 15193456 Review.
Cited by
- Novel mechanism of nuclear receptor corepressor interaction dictated by activation function 2 helix determinants.
Moraitis AN, Giguère V, Thompson CC. Moraitis AN, et al. Mol Cell Biol. 2002 Oct;22(19):6831-41. doi: 10.1128/MCB.22.19.6831-6841.2002. Mol Cell Biol. 2002. PMID: 12215540 Free PMC article. - A natural allele of Nxf1 suppresses retrovirus insertional mutations.
Floyd JA, Gold DA, Concepcion D, Poon TH, Wang X, Keithley E, Chen D, Ward EJ, Chinn SB, Friedman RA, Yu HT, Moriwaki K, Shiroishi T, Hamilton BA. Floyd JA, et al. Nat Genet. 2003 Nov;35(3):221-8. doi: 10.1038/ng1247. Epub 2003 Sep 28. Nat Genet. 2003. PMID: 14517553 Free PMC article. - Hairless regulates heterochromatin maintenance and muscle stem cell function as a histone demethylase antagonist.
Liu L, Rodriguez-Mateo C, Huang P, Huang A, Lieu A, Mao M, Chung M, Yang S, Yu K, Charville GW, Gan Q, Rando TA. Liu L, et al. Proc Natl Acad Sci U S A. 2021 Sep 14;118(37):e2025281118. doi: 10.1073/pnas.2025281118. Proc Natl Acad Sci U S A. 2021. PMID: 34493660 Free PMC article. - Molecular evolution of HR, a gene that regulates the postnatal cycle of the hair follicle.
Abbasi AA. Abbasi AA. Sci Rep. 2011;1:32. doi: 10.1038/srep00032. Epub 2011 Jul 6. Sci Rep. 2011. PMID: 22355551 Free PMC article. - Poly(rC) binding protein 2 acts as a negative regulator of IRES-mediated translation of Hr mRNA.
Kim JK, Kim I, Choi K, Choi JH, Kim E, Lee HY, Park J, Kim Yoon S. Kim JK, et al. Exp Mol Med. 2018 Feb 9;50(2):e441. doi: 10.1038/emm.2017.262. Exp Mol Med. 2018. PMID: 29422543 Free PMC article.
References
- Ahmad W, Faiyaz ul Haque M, Brancolini V, Tsou HC, ul Haque S, Lam H, Aita VM, Owen J, deBlaquiere M, Frank J, et al. Alopecia universalis associated with a mutation in the human hairless gene. Science. 1998;279:720–724. - PubMed
- Ahmad W, Panteleyev AA, Christiano AM. The molecular basis of congenital atrichia in humans and mice: Mutations in the hairless gene. J Investig Dermatol Symp Proc. 1999;4:240–243. - PubMed
- Aita VM, Ahmad W, Panteleyev AA, Kozlowska U, Kozlowska A, Gilliam TC, Jablonska S, Christiano AM. A novel missense mutation (C622G) in the zinc-finger domain of the human hairless gene associated with congenital atrichia with papular lesions. Exp Dermatol. 2000;9:157–162. - PubMed
- Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho RA. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials