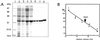A PP2C-type phosphatase dephosphorylates the PII signaling protein in the cyanobacterium Synechocystis PCC 6803 - PubMed (original) (raw)
A PP2C-type phosphatase dephosphorylates the PII signaling protein in the cyanobacterium Synechocystis PCC 6803
A Irmler et al. Proc Natl Acad Sci U S A. 2001.
Abstract
The family of the PII signal transduction proteins contains the most highly conserved signaling proteins in nature. The cyanobacterial PII-homologue transmits signals of the cellular nitrogen status and carbon status through phosphorylation of a seryl-residue. To identify the enzyme responsible for dephosphorylation of the phosphorylated PII protein in Synechocystis PCC 6803, prospective phosphatase encoding genes were inactivated by targeted insertion of kanamycin resistance cassettes. Disruption of ORF sll1771 generates a mutant unable to dephosphorylate PII under various experimental conditions. On the basis of conserved signature motifs, the sll1771 product (termed PphA) is a member of the protein phosphatase 2C (PP2C) superfamily, which is characterized by Mg(2+)/Mn(2+)-dependent catalytic activity. Biochemical analysis of overexpressed and purified PphA confirms its PP2C-type enzymatic properties and demonstrated its reactivity toward the phosphorylated PII protein. Thus, PphA is the first protein phosphatase in Synechocystis PCC 6803 for which the physiological substrate and function is known.
Figures
Figure 1
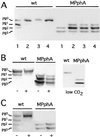
Phosphorylation state of PII in wild-type (wt) and sll1771 inactivated (MPphA) cells of Synechocystis PCC6803. (A) Response to ammonium treatment; cells were first grown in BG11-medium (lane 1), then NH4Cl was added (5 mM final concentration) and after 5 min (2), 15 min (3), and 30 min (4) aliquots were removed and the phosphorylation state of PII was analyzed. (B) Response to different carbon conditions: (Left) Cells grown in BG11-medium were treated with an inhibitor of CO2-fixation,
d
,
l
-glyceraldehyde (
d
,
l
-GA). Negative control (−) and after 5 min in the presence of 30 mM
d
,
l
-GA (+); (Right) cells grown under CO2-poor conditions in Allan and Arnon's medium, which is devoid of carbonate and bicarbonate. (C) Response toward methylviologen (MV): to BG11-grown cells 0.1 mM MV was added and, after 5 min, the phosphorylation state of PII was analyzed.
Figure 2
(A) Organization of the DNA region containing sll1770, sll1771, and sll1772 according to the gene map from the Cyanobase database (1) and position of the inserted kanamycin resistance cassette (KmR) in mutants of sll1770 and sll1771. Mutants in sll1771 were generated with the KmR cassette in both orientations, indicated by the orientation of the arrows. The bent arrow indicates the putative transcriptional start site, as deduced from 5′ RACE assay. (B) Northern blot analysis of_pphA_-transcripts from Synechocystis PCC 6803 (lane 1) and the sll1771 mutants with the KmR cassette in the same orientation (lane 2) or opposite (lane 3) to_pphA_. RNA was prepared from nitrate-grown cells and was probed with the pphA gene.
Figure 3
Phylogenetic analysis of PphA and the prospective serine/threonine protein phosphatases of Synechocystis PCC 6803 together with selected PP2C members from bacterial and eukaryotic origin. The predicted size (amino acids) of the proteins is given in brackets. Sequence homology searches were carried out by using the BLAST analysis (37). The dendrogram was constructed with the programs PHYLIP Version 3.572 and CLUSTALX (38). Accession nos. of the sequences (from top to bottom): BAB08417; P35813; BAA10651; BAA18661; L35574; BAA17283; P37979; BAA18228; P37475; BAA10367; CAB65437; BAA16771; AAF73659; E71538; AF223364; BAB06224; G69878; CAB60748; CAA10712; RAN02136; RNPU03387; BAA17671; H75265; A83438.
Figure 4
(A) Purification of overproduced PphA shown by Coomassie-blue-stained 12.5% SDS/PAGE. The lanes correspond to the different purification steps as described in Materials and Methods. 1, molecular weight marker; 2, S10; 3, S100; 4, first (NH4)2SO4 precipitate; 5, DEAE Sepharose Fast Flow-pool; 6, UnoQ-pool; 7, Methyl HIC-pool; 8, Superdex 200-pool. (B) Determination of the native molecular size of PphA by gel-permeation chromatography. PphA and proteins used as size standards (1, alcohol dehydrogenase, 150 kDa; 2, BSA, 67 kDa; 3, ovalbumin, 43 kDa; 4, carbonic anhydrase, 29 kDa; 5, cytochrome C, 12.4 kDa) were passed separately through a Superdex S200 h 10/30 column (Amersham Pharmacia) equilibrated in 20 mM Tris⋅Cl (pH 7.4), 150 mM NaCl, 5 mM MgCl2, 0.5 mM EDTA, 2 mM DTT, 1 mM benzamidine, and 1 mM ɛ-aminocaproic acid.
Figure 5
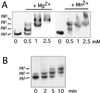
In vitro reactivity of PphA toward purified phosphorylated PII. (A) Mg2+/Mn2+ dependence of PII-P dephosphorylation. Mg2+/Mn2+-free PphA (10 ng) was incubated with PII-P in the presence of 0, 0.5, 1, or 2.5 mM MgCl2 or MnCl2 as indicated. After 30 min the phosphorylation state of PII was determined. (B) Time course of in vitro PII-P dephosphorylation. Standard reactions (with 10 mM MgCl2) were started by the addition of 3 ng PphA. After 2, 5, and 10 min the reactions were stopped and analyzed.
Similar articles
- The novel protein phosphatase PphA from Synechocystis PCC 6803 controls dephosphorylation of the signalling protein PII.
Ruppert U, Irmler A, Kloft N, Forchhammer K. Ruppert U, et al. Mol Microbiol. 2002 May;44(3):855-64. doi: 10.1046/j.1365-2958.2002.02927.x. Mol Microbiol. 2002. PMID: 11994164 - Protein phosphatase PphA from Synechocystis sp. PCC 6803: the physiological framework of PII-P dephosphorylation.
Kloft N, Rasch G, Forchhammer K. Kloft N, et al. Microbiology (Reading). 2005 Apr;151(Pt 4):1275-83. doi: 10.1099/mic.0.27771-0. Microbiology (Reading). 2005. PMID: 15817794 - Determinants for substrate specificity of the bacterial PP2C protein phosphatase tPphA from Thermosynechococcus elongatus.
Su J, Forchhammer K. Su J, et al. FEBS J. 2013 Jan;280(2):694-707. doi: 10.1111/j.1742-4658.2011.08466.x. Epub 2012 Jan 23. FEBS J. 2013. PMID: 22212593 - Global carbon/nitrogen control by PII signal transduction in cyanobacteria: from signals to targets.
Forchhammer K. Forchhammer K. FEMS Microbiol Rev. 2004 Jun;28(3):319-33. doi: 10.1016/j.femsre.2003.11.001. FEMS Microbiol Rev. 2004. PMID: 15449606 Review. - Keeping in touch with PII: PII-interacting proteins in unicellular cyanobacteria.
Osanai T, Tanaka K. Osanai T, et al. Plant Cell Physiol. 2007 Jul;48(7):908-14. doi: 10.1093/pcp/pcm072. Epub 2007 Jun 12. Plant Cell Physiol. 2007. PMID: 17566056 Review.
Cited by
- Mycobacterial serine/threonine phosphatase PstP is phosphoregulated and localized to mediate control of cell wall metabolism.
Shamma F, Rego EH, Boutte CC. Shamma F, et al. Mol Microbiol. 2022 Jul;118(1-2):47-60. doi: 10.1111/mmi.14951. Epub 2022 Jun 20. Mol Microbiol. 2022. PMID: 35670057 Free PMC article. - A low molecular weight protein tyrosine phosphatase from Synechocystis sp. strain PCC 6803: enzymatic characterization and identification of its potential substrates.
Mukhopadhyay A, Kennelly PJ. Mukhopadhyay A, et al. J Biochem. 2011 May;149(5):551-62. doi: 10.1093/jb/mvr014. Epub 2011 Feb 1. J Biochem. 2011. PMID: 21288886 Free PMC article. - Identification of Rhodospirillum rubrum GlnB variants that are altered in their ability to interact with different targets in response to nitrogen status signals.
Zhu Y, Conrad MC, Zhang Y, Roberts GP. Zhu Y, et al. J Bacteriol. 2006 Mar;188(5):1866-74. doi: 10.1128/JB.188.5.1866-1874.2006. J Bacteriol. 2006. PMID: 16484197 Free PMC article. - Phosphoproteome of the cyanobacterium Synechocystis sp. PCC 6803 and its dynamics during nitrogen starvation.
Spät P, Maček B, Forchhammer K. Spät P, et al. Front Microbiol. 2015 Mar 31;6:248. doi: 10.3389/fmicb.2015.00248. eCollection 2015. Front Microbiol. 2015. PMID: 25873915 Free PMC article. - Post-translational modification of P II signal transduction proteins.
Merrick M. Merrick M. Front Microbiol. 2015 Jan 6;5:763. doi: 10.3389/fmicb.2014.00763. eCollection 2014. Front Microbiol. 2015. PMID: 25610437 Free PMC article. Review.
References
- Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, et al. DNA Res. 1996;3:109–136. - PubMed
- Zhang C C. Mol Microbiol. 1996;20:9–15. - PubMed
- Shi L, Potts M, Kenelly M. FEMS Microbiol Rev. 1998;22:229–253. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases