Common origin of four diverse families of large eukaryotic DNA viruses - PubMed (original) (raw)
Comparative Study
Common origin of four diverse families of large eukaryotic DNA viruses
L M Iyer et al. J Virol. 2001 Dec.
Abstract
Comparative analysis of the protein sequences encoded in the genomes of three families of large DNA viruses that replicate, completely or partly, in the cytoplasm of eukaryotic cells (poxviruses, asfarviruses, and iridoviruses) and phycodnaviruses that replicate in the nucleus reveals 9 genes that are shared by all of these viruses and 22 more genes that are present in at least three of the four compared viral families. Although orthologous proteins from different viral families typically show weak sequence similarity, because of which some of them have not been identified previously, at least five of the conserved genes appear to be synapomorphies (shared derived characters) that unite these four viral families, to the exclusion of all other known viruses and cellular life forms. Cladistic analysis with the genes shared by at least two viral families as evolutionary characters supports the monophyly of poxviruses, asfarviruses, iridoviruses, and phycodnaviruses. The results of genome comparison allow a tentative reconstruction of the ancestral viral genome and suggest that the common ancestor of all of these viral families was a nucleocytoplasmic virus with an icosahedral capsid, which encoded complex systems for DNA replication and transcription, a redox protein involved in disulfide bond formation in virion membrane proteins, and probably inhibitors of apoptosis. The conservation of the disulfide-oxidoreductase, a major capsid protein, and two virion membrane proteins indicates that the odd-shaped virions of poxviruses have evolved from the more common icosahedral virion seen in asfarviruses, iridoviruses, and phycodnaviruses.
Figures
FIG. 1
Multiple alignments of conserved proteins that define the cytoplasmic DNA virus clade. (A) D5R-like helicases. With the PBCV ATPase as the seed, the ESV ortholog and many phage primases were recovered with highly significant Expectation (E) values in the first iteration. Proteins from the other NCLDV and the distantly related papillomavirus, parvovirus, and positive-strand RNA viruses were recovered in the second and third iterations with E-values of <10−3. For example, ASFV C962R was recovered with an E-value of 10−8 in the third iteration. Further transitive searches identified all of the members of superfamily III helicase. (B) A32L-like ATPases. With the PBCV ATPase as the seed, iridoviral orthologs were recovered in the first iteration with an E-value of <10−5. Orthologs from all other NCLDV were recovered by the third iteration with significant E-values such as 3 × 10−19 for MCV and 2 × 10−04 for ASFV orthologs. (C) A1L-like transcription factors. A profile made with previously detected FCS domains from the polyhomeotic and FIM families of proteins, when run against the NCLDV protein sets, with an inclusion cutoff of 0.01, recovered all members of this family; VV A1L, for example, was recovered with an E-value of 10−4. (D) D13L-like capsid proteins. With p50 of the Spodoptera exigua ascovirus as the seed, the PBCV and other iridoviral capsid proteins were recovered with E-values of <2 × 10−8. The ASFV ortholog was detected in the third iteration with an E-value of 3 × 10−3, and the poxviral D13L-like proteins were recovered at borderline E-values (0.14) in the fourth iteration. When a profile made from the alignment of the PBCV, iridovirus, and ASFV sequences was run against a database of all NCLDV proteins, the poxviral orthologs were detected as top hits, with E-values of <10−5. The probability of the conserved motifs shown here to occur in these proteins by chance was <10−15, as computed by using the MACAW program (49). (E) L1R/F9L-like virion membrane proteins. With CIV 048L as the seed, the ASFV and PBCV orthologs were recovered in the second iteration, with E-values of 8 × 10−4 and 10−3, respectively. The entomopoxviral orthologs were detected in the third iteration with an E-value of 2 × 10−4. A transitive search with the entomopoxviral proteins recovered the other poxviral proteins with E-values of <10−3. Each protein is denoted by the corresponding gene name followed by species abbreviation and the GenBank Identifier (GI) number. The numbers preceding and following the alignments indicate the positions of the first and last residues of the aligned regions in the corresponding protein sequences. The numbers between aligned blocks indicate the number of inserted residues that were omitted from the figure. The coloring reflects the conservation of amino acid residues at 85% consensus. The coloring scheme and the consensus abbreviations are as follows: hydrophobic residues (LIYFMWACV) are designated “h” in the consensus line, aliphatic (LIAV) residues are also shaded yellow and designated “l,” alcohol (S,T) is blue and designated “o,” charged (KERDH) residues are purple and designated “c,” polar (STEDRKHNQ) residues are purple and designated “p,” small (SACGDNPVT) residues are green and designated “s,” big (LIFMWYERKQ) residues are shaded gray and designated “b.” Conserved cysteines predicted to form a Zn-finger structure (C) or a disulfide bond (E) are indicated by white letters against a red background. Secondary structure elements predicted by using the PHD program are indicated in panels C and D; where “E” indicates extended conformation (b-strand) and “H” indicates the α-helix. The abbreviations for the NCLDV are defined in Materials and Methods. Additional abbreviations: AAV, adeno-associated virus 5; AcNPV, Autographa californica nucleopolyhedrovirus; Bf, Bacteroides fragilis, Ce, Caenorhabditis elegans; Cglu, Corynebacterium glutamicum; Cpf, Clostridium perfringens; Dm, Drosophila melanogaster; DpAV4, Diadromus pulchellu_s ascovirus; Ec, Escherichia coli; HPV08, human papillomavirus type 8; Hs, Homo sapiens; LcbA2, Lactobacillus casei bacteriophage A2; Mace, Methanosarcina acetivorans; MStV, maize streak virus; phi-105, Bacteriophage phi-105; phiC31, Bacteriophage phiC31; Polio, human poliovirus 1; SacV, Spodoptera exigua ascovirus; Si, Sulfolobus islandicus; SV40, Simian virus 40; Xf, Xylella fastidiosa._
FIG. 1
Multiple alignments of conserved proteins that define the cytoplasmic DNA virus clade. (A) D5R-like helicases. With the PBCV ATPase as the seed, the ESV ortholog and many phage primases were recovered with highly significant Expectation (E) values in the first iteration. Proteins from the other NCLDV and the distantly related papillomavirus, parvovirus, and positive-strand RNA viruses were recovered in the second and third iterations with E-values of <10−3. For example, ASFV C962R was recovered with an E-value of 10−8 in the third iteration. Further transitive searches identified all of the members of superfamily III helicase. (B) A32L-like ATPases. With the PBCV ATPase as the seed, iridoviral orthologs were recovered in the first iteration with an E-value of <10−5. Orthologs from all other NCLDV were recovered by the third iteration with significant E-values such as 3 × 10−19 for MCV and 2 × 10−04 for ASFV orthologs. (C) A1L-like transcription factors. A profile made with previously detected FCS domains from the polyhomeotic and FIM families of proteins, when run against the NCLDV protein sets, with an inclusion cutoff of 0.01, recovered all members of this family; VV A1L, for example, was recovered with an E-value of 10−4. (D) D13L-like capsid proteins. With p50 of the Spodoptera exigua ascovirus as the seed, the PBCV and other iridoviral capsid proteins were recovered with E-values of <2 × 10−8. The ASFV ortholog was detected in the third iteration with an E-value of 3 × 10−3, and the poxviral D13L-like proteins were recovered at borderline E-values (0.14) in the fourth iteration. When a profile made from the alignment of the PBCV, iridovirus, and ASFV sequences was run against a database of all NCLDV proteins, the poxviral orthologs were detected as top hits, with E-values of <10−5. The probability of the conserved motifs shown here to occur in these proteins by chance was <10−15, as computed by using the MACAW program (49). (E) L1R/F9L-like virion membrane proteins. With CIV 048L as the seed, the ASFV and PBCV orthologs were recovered in the second iteration, with E-values of 8 × 10−4 and 10−3, respectively. The entomopoxviral orthologs were detected in the third iteration with an E-value of 2 × 10−4. A transitive search with the entomopoxviral proteins recovered the other poxviral proteins with E-values of <10−3. Each protein is denoted by the corresponding gene name followed by species abbreviation and the GenBank Identifier (GI) number. The numbers preceding and following the alignments indicate the positions of the first and last residues of the aligned regions in the corresponding protein sequences. The numbers between aligned blocks indicate the number of inserted residues that were omitted from the figure. The coloring reflects the conservation of amino acid residues at 85% consensus. The coloring scheme and the consensus abbreviations are as follows: hydrophobic residues (LIYFMWACV) are designated “h” in the consensus line, aliphatic (LIAV) residues are also shaded yellow and designated “l,” alcohol (S,T) is blue and designated “o,” charged (KERDH) residues are purple and designated “c,” polar (STEDRKHNQ) residues are purple and designated “p,” small (SACGDNPVT) residues are green and designated “s,” big (LIFMWYERKQ) residues are shaded gray and designated “b.” Conserved cysteines predicted to form a Zn-finger structure (C) or a disulfide bond (E) are indicated by white letters against a red background. Secondary structure elements predicted by using the PHD program are indicated in panels C and D; where “E” indicates extended conformation (b-strand) and “H” indicates the α-helix. The abbreviations for the NCLDV are defined in Materials and Methods. Additional abbreviations: AAV, adeno-associated virus 5; AcNPV, Autographa californica nucleopolyhedrovirus; Bf, Bacteroides fragilis, Ce, Caenorhabditis elegans; Cglu, Corynebacterium glutamicum; Cpf, Clostridium perfringens; Dm, Drosophila melanogaster; DpAV4, Diadromus pulchellu_s ascovirus; Ec, Escherichia coli; HPV08, human papillomavirus type 8; Hs, Homo sapiens; LcbA2, Lactobacillus casei bacteriophage A2; Mace, Methanosarcina acetivorans; MStV, maize streak virus; phi-105, Bacteriophage phi-105; phiC31, Bacteriophage phiC31; Polio, human poliovirus 1; SacV, Spodoptera exigua ascovirus; Si, Sulfolobus islandicus; SV40, Simian virus 40; Xf, Xylella fastidiosa._
FIG. 1
Multiple alignments of conserved proteins that define the cytoplasmic DNA virus clade. (A) D5R-like helicases. With the PBCV ATPase as the seed, the ESV ortholog and many phage primases were recovered with highly significant Expectation (E) values in the first iteration. Proteins from the other NCLDV and the distantly related papillomavirus, parvovirus, and positive-strand RNA viruses were recovered in the second and third iterations with E-values of <10−3. For example, ASFV C962R was recovered with an E-value of 10−8 in the third iteration. Further transitive searches identified all of the members of superfamily III helicase. (B) A32L-like ATPases. With the PBCV ATPase as the seed, iridoviral orthologs were recovered in the first iteration with an E-value of <10−5. Orthologs from all other NCLDV were recovered by the third iteration with significant E-values such as 3 × 10−19 for MCV and 2 × 10−04 for ASFV orthologs. (C) A1L-like transcription factors. A profile made with previously detected FCS domains from the polyhomeotic and FIM families of proteins, when run against the NCLDV protein sets, with an inclusion cutoff of 0.01, recovered all members of this family; VV A1L, for example, was recovered with an E-value of 10−4. (D) D13L-like capsid proteins. With p50 of the Spodoptera exigua ascovirus as the seed, the PBCV and other iridoviral capsid proteins were recovered with E-values of <2 × 10−8. The ASFV ortholog was detected in the third iteration with an E-value of 3 × 10−3, and the poxviral D13L-like proteins were recovered at borderline E-values (0.14) in the fourth iteration. When a profile made from the alignment of the PBCV, iridovirus, and ASFV sequences was run against a database of all NCLDV proteins, the poxviral orthologs were detected as top hits, with E-values of <10−5. The probability of the conserved motifs shown here to occur in these proteins by chance was <10−15, as computed by using the MACAW program (49). (E) L1R/F9L-like virion membrane proteins. With CIV 048L as the seed, the ASFV and PBCV orthologs were recovered in the second iteration, with E-values of 8 × 10−4 and 10−3, respectively. The entomopoxviral orthologs were detected in the third iteration with an E-value of 2 × 10−4. A transitive search with the entomopoxviral proteins recovered the other poxviral proteins with E-values of <10−3. Each protein is denoted by the corresponding gene name followed by species abbreviation and the GenBank Identifier (GI) number. The numbers preceding and following the alignments indicate the positions of the first and last residues of the aligned regions in the corresponding protein sequences. The numbers between aligned blocks indicate the number of inserted residues that were omitted from the figure. The coloring reflects the conservation of amino acid residues at 85% consensus. The coloring scheme and the consensus abbreviations are as follows: hydrophobic residues (LIYFMWACV) are designated “h” in the consensus line, aliphatic (LIAV) residues are also shaded yellow and designated “l,” alcohol (S,T) is blue and designated “o,” charged (KERDH) residues are purple and designated “c,” polar (STEDRKHNQ) residues are purple and designated “p,” small (SACGDNPVT) residues are green and designated “s,” big (LIFMWYERKQ) residues are shaded gray and designated “b.” Conserved cysteines predicted to form a Zn-finger structure (C) or a disulfide bond (E) are indicated by white letters against a red background. Secondary structure elements predicted by using the PHD program are indicated in panels C and D; where “E” indicates extended conformation (b-strand) and “H” indicates the α-helix. The abbreviations for the NCLDV are defined in Materials and Methods. Additional abbreviations: AAV, adeno-associated virus 5; AcNPV, Autographa californica nucleopolyhedrovirus; Bf, Bacteroides fragilis, Ce, Caenorhabditis elegans; Cglu, Corynebacterium glutamicum; Cpf, Clostridium perfringens; Dm, Drosophila melanogaster; DpAV4, Diadromus pulchellu_s ascovirus; Ec, Escherichia coli; HPV08, human papillomavirus type 8; Hs, Homo sapiens; LcbA2, Lactobacillus casei bacteriophage A2; Mace, Methanosarcina acetivorans; MStV, maize streak virus; phi-105, Bacteriophage phi-105; phiC31, Bacteriophage phiC31; Polio, human poliovirus 1; SacV, Spodoptera exigua ascovirus; Si, Sulfolobus islandicus; SV40, Simian virus 40; Xf, Xylella fastidiosa._
FIG. 1
Multiple alignments of conserved proteins that define the cytoplasmic DNA virus clade. (A) D5R-like helicases. With the PBCV ATPase as the seed, the ESV ortholog and many phage primases were recovered with highly significant Expectation (E) values in the first iteration. Proteins from the other NCLDV and the distantly related papillomavirus, parvovirus, and positive-strand RNA viruses were recovered in the second and third iterations with E-values of <10−3. For example, ASFV C962R was recovered with an E-value of 10−8 in the third iteration. Further transitive searches identified all of the members of superfamily III helicase. (B) A32L-like ATPases. With the PBCV ATPase as the seed, iridoviral orthologs were recovered in the first iteration with an E-value of <10−5. Orthologs from all other NCLDV were recovered by the third iteration with significant E-values such as 3 × 10−19 for MCV and 2 × 10−04 for ASFV orthologs. (C) A1L-like transcription factors. A profile made with previously detected FCS domains from the polyhomeotic and FIM families of proteins, when run against the NCLDV protein sets, with an inclusion cutoff of 0.01, recovered all members of this family; VV A1L, for example, was recovered with an E-value of 10−4. (D) D13L-like capsid proteins. With p50 of the Spodoptera exigua ascovirus as the seed, the PBCV and other iridoviral capsid proteins were recovered with E-values of <2 × 10−8. The ASFV ortholog was detected in the third iteration with an E-value of 3 × 10−3, and the poxviral D13L-like proteins were recovered at borderline E-values (0.14) in the fourth iteration. When a profile made from the alignment of the PBCV, iridovirus, and ASFV sequences was run against a database of all NCLDV proteins, the poxviral orthologs were detected as top hits, with E-values of <10−5. The probability of the conserved motifs shown here to occur in these proteins by chance was <10−15, as computed by using the MACAW program (49). (E) L1R/F9L-like virion membrane proteins. With CIV 048L as the seed, the ASFV and PBCV orthologs were recovered in the second iteration, with E-values of 8 × 10−4 and 10−3, respectively. The entomopoxviral orthologs were detected in the third iteration with an E-value of 2 × 10−4. A transitive search with the entomopoxviral proteins recovered the other poxviral proteins with E-values of <10−3. Each protein is denoted by the corresponding gene name followed by species abbreviation and the GenBank Identifier (GI) number. The numbers preceding and following the alignments indicate the positions of the first and last residues of the aligned regions in the corresponding protein sequences. The numbers between aligned blocks indicate the number of inserted residues that were omitted from the figure. The coloring reflects the conservation of amino acid residues at 85% consensus. The coloring scheme and the consensus abbreviations are as follows: hydrophobic residues (LIYFMWACV) are designated “h” in the consensus line, aliphatic (LIAV) residues are also shaded yellow and designated “l,” alcohol (S,T) is blue and designated “o,” charged (KERDH) residues are purple and designated “c,” polar (STEDRKHNQ) residues are purple and designated “p,” small (SACGDNPVT) residues are green and designated “s,” big (LIFMWYERKQ) residues are shaded gray and designated “b.” Conserved cysteines predicted to form a Zn-finger structure (C) or a disulfide bond (E) are indicated by white letters against a red background. Secondary structure elements predicted by using the PHD program are indicated in panels C and D; where “E” indicates extended conformation (b-strand) and “H” indicates the α-helix. The abbreviations for the NCLDV are defined in Materials and Methods. Additional abbreviations: AAV, adeno-associated virus 5; AcNPV, Autographa californica nucleopolyhedrovirus; Bf, Bacteroides fragilis, Ce, Caenorhabditis elegans; Cglu, Corynebacterium glutamicum; Cpf, Clostridium perfringens; Dm, Drosophila melanogaster; DpAV4, Diadromus pulchellu_s ascovirus; Ec, Escherichia coli; HPV08, human papillomavirus type 8; Hs, Homo sapiens; LcbA2, Lactobacillus casei bacteriophage A2; Mace, Methanosarcina acetivorans; MStV, maize streak virus; phi-105, Bacteriophage phi-105; phiC31, Bacteriophage phiC31; Polio, human poliovirus 1; SacV, Spodoptera exigua ascovirus; Si, Sulfolobus islandicus; SV40, Simian virus 40; Xf, Xylella fastidiosa._
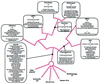
FIG. 2
Consensus cladogram of cytoplasmic DNA viruses. The cladistic analysis was performed as described in the text. The proteins that were probably present in the common ancestor of the universally supported NCLDV clade are superimposed on the consensus tree. Also shown on the consensus tree are the state changes in each of the terminal lineages and the strictly supported clades. The plus sign indicates a character that is most parsimoniously explained as an independent gain that was most likely acquired through horizontal transfer between the viral genome or through transfer from the host genome. The minus sign denotes the loss of an ancestral character in a particular lineage.
Similar articles
- Evolutionary genomics of nucleo-cytoplasmic large DNA viruses.
Iyer LM, Balaji S, Koonin EV, Aravind L. Iyer LM, et al. Virus Res. 2006 Apr;117(1):156-84. doi: 10.1016/j.virusres.2006.01.009. Epub 2006 Feb 21. Virus Res. 2006. PMID: 16494962 Review. - Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences.
Koonin EV, Dolja VV. Koonin EV, et al. Crit Rev Biochem Mol Biol. 1993;28(5):375-430. doi: 10.3109/10409239309078440. Crit Rev Biochem Mol Biol. 1993. PMID: 8269709 Review. - [The great virus comeback].
Forterre P. Forterre P. Biol Aujourdhui. 2013;207(3):153-68. doi: 10.1051/jbio/2013018. Epub 2013 Dec 13. Biol Aujourdhui. 2013. PMID: 24330969 Review. French. - The genome of molluscum contagiosum virus: analysis and comparison with other poxviruses.
Senkevich TG, Koonin EV, Bugert JJ, Darai G, Moss B. Senkevich TG, et al. Virology. 1997 Jun 23;233(1):19-42. doi: 10.1006/viro.1997.8607. Virology. 1997. PMID: 9201214 - Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging.
Iyer LM, Makarova KS, Koonin EV, Aravind L. Iyer LM, et al. Nucleic Acids Res. 2004 Oct 5;32(17):5260-79. doi: 10.1093/nar/gkh828. Print 2004. Nucleic Acids Res. 2004. PMID: 15466593 Free PMC article.
Cited by
- Current capsid assembly models of icosahedral nucleocytoviricota viruses.
Xian Y, Xiao C. Xian Y, et al. Adv Virus Res. 2020;108:275-313. doi: 10.1016/bs.aivir.2020.09.006. Epub 2020 Oct 5. Adv Virus Res. 2020. PMID: 33837719 Free PMC article. Review. - Giant Viruses of Amoebas: An Update.
Aherfi S, Colson P, La Scola B, Raoult D. Aherfi S, et al. Front Microbiol. 2016 Mar 22;7:349. doi: 10.3389/fmicb.2016.00349. eCollection 2016. Front Microbiol. 2016. PMID: 27047465 Free PMC article. Review. - Related giant viruses in distant locations and different habitats: Acanthamoeba polyphaga moumouvirus represents a third lineage of the Mimiviridae that is close to the megavirus lineage.
Yoosuf N, Yutin N, Colson P, Shabalina SA, Pagnier I, Robert C, Azza S, Klose T, Wong J, Rossmann MG, La Scola B, Raoult D, Koonin EV. Yoosuf N, et al. Genome Biol Evol. 2012;4(12):1324-30. doi: 10.1093/gbe/evs109. Genome Biol Evol. 2012. PMID: 23221609 Free PMC article. - Packing and trimer-to-dimer protein reconstruction in icosahedral viral shells with a single type of symmetrical structural unit.
Rochal SB, Konevtsova OV, Roshal DS, Božič A, Golushko IY, Podgornik R. Rochal SB, et al. Nanoscale Adv. 2022 Sep 21;4(21):4677-4688. doi: 10.1039/d2na00461e. eCollection 2022 Oct 25. Nanoscale Adv. 2022. PMID: 36341291 Free PMC article. - The Vaccinia Virus DNA Helicase Structure from Combined Single-Particle Cryo-Electron Microscopy and AlphaFold2 Prediction.
Hutin S, Ling WL, Tarbouriech N, Schoehn G, Grimm C, Fischer U, Burmeister WP. Hutin S, et al. Viruses. 2022 Oct 7;14(10):2206. doi: 10.3390/v14102206. Viruses. 2022. PMID: 36298761 Free PMC article.
References
- Altschul S F, Koonin E V. PSI-BLAST—a tool for making discoveries in sequence databases. Trends Biochem Sci. 1998;23:444–447. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources