Functional cooperation of Epstein-Barr virus nuclear antigen 2 and the survival motor neuron protein in transactivation of the viral LMP1 promoter - PubMed (original) (raw)
Functional cooperation of Epstein-Barr virus nuclear antigen 2 and the survival motor neuron protein in transactivation of the viral LMP1 promoter
M D Voss et al. J Virol. 2001 Dec.
Abstract
Epstein-Barr virus nuclear antigen 2 (EBNA2) is essential for viral transformation of B cells and transactivates cellular and viral target genes by binding RBPJkappa tethered to cognate promoter elements. EBNA2 interacts with the DEAD-box protein DP103 (DDX20/Gemin3), which in turn is complexed to the survival motor neuron (SMN) protein. SMN is implicated in RNA processing, but a role in transcriptional regulation has also been suggested. Here, we show that DP103 and SMN are complexed in B cells and that SMN coactivates the viral LMP promoter in the presence of EBNA2 in reporter gene assays and in vivo. Subcellular localization studies revealed that nuclear gems and/or coiled bodies containing DP103 and SMN are targeted by EBNA2. Protein-protein interaction experiments demonstrated that DP103 binds to SMN exon 6 and that both EBNA2 and SMN interact with the C terminus of DP103. Furthermore, a DP103 binding-deficient SMN mutant was released from nuclear gems and/or coiled bodies and further enhanced coactivation. In addition, impaired transactivation of a DP103 binding-deficient EBNA2 mutant was rescued by overexpression of SMN. Testing different promoter constructs in luciferase assays showed that RBPJkappa is required but not sufficient for coactivation by EBNA2 and SMN. Overall, our data suggest that EBNA2 might target spliceosomal complexes by binding to DP103, thereby releasing SMN which subsequently exerts a coactivational function within the RNA-polymerase II transcription complex on the LMP1 promoter.
Figures
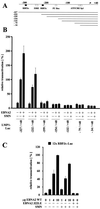
FIG. 1
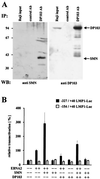
(A) Interaction of DP103 and SMN in B lymphocytes. Coimmunoprecipitations (IP) from Raji cell extracts were performed with DP103-specific MAb 9A3 (IP: DP103 Ab) or an irrelevant control antibody (anti-TrypE 3A6, IP: control Ab), followed by SDS–10% PAGE and Western blotting. Precipitated proteins were detected with anti-SMN-N serum (Santa Cruz Biochemicals) (left panel, WB: anti SMN) or anti-DP103 MAb 8H4 (right panel, WB: anti DP103). The positions of SMN and DP103 are indicated by arrows. Lanes designated Raji input represent ca. 1% of unprecipitated Raji cell extract. The positions of the molecular mass marker proteins are indicated on the left side (in kilodaltons). (B) SMN coactivates the viral LMP1 promoter in the presence of EBNA2. BJAB cells were transfected with luciferase reporter constructs encoding positions −327/+40 (EBNA2 responsive) or −154/+40 (nonresponsive) of the LMP1 promoter (4 μg) and the indicated combinations of pSG5 constructs encoding EBNA2 or HA-tagged SMN and DP103 (10 μg). After 48 h, the cells were lysed by freeze-thawing, and the luciferase activity was measured. The transfection efficiency was determined by scanning the expression of cotransfected pEGFP-C1 vector (2 μg) by FACS analysis prior to lysis of the cells. For each experiment, luciferase values standardized for transfection efficiency were calculated relative to the values obtained by EBNA2 and the respective full-length promoter construct (set to 100%). Graphs represent the mean values of five independent experiments (± the standard error of the mean [SEM]).
FIG. 2
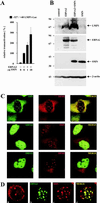
(A) Dose-dependent coactivation of the −327/+40 LMP1 promoter by coexpression of EBNA2 and increasing amounts of HA-tagged SMN (indicated in micrograms). Assays were performed as described for Fig. 1B. Graphs represent the mean values of three independent experiments performed in duplicate (±SEM). (B) SMN increases EBNA2-mediated induction of endogenous LMP1 protein. EBV-positive P3HR1 cells were transfected with pSG5 constructs (15 μg) encoding EBNA2 or HA-tagged SMN, as indicated. Cells were harvested and subjected to SDS–10% PAGE and Western blotting by using MAbs S12 (anti-LMP1), R3 (anti-EBNA2), 3F10 (anti-HA), and anti-β-actin MAb (Sigma). The positions of the respective proteins are indicated by arrows. The positions of the molecular mass markers (in kilodaltons) are indicated on the left side. (C) Subcellular distribution of EBNA2, DP103, and SMN. HeLa cells transfected with pSG5-HA (red signals) or pEGFP-C1 (green signals) constructs (5 μg) encoding the corresponding fusion proteins of EBNA2, DP103, or SMN were immunostained and analyzed by confocal laser scanning microscopy. HA-tagged proteins were visualized by using anti-HA 3F10/anti-rat TRITC MAbs. The localizations of coexpressed SMN and DP103 (upper panel), EBNA2 and SMN (middle panel), or EBNA2 and DP103 (lower panel) are shown. In the merged images, colocalization results in a yellow signal. (D) Subcellular localization of endogenous SMN and EGFP-EBNA2 in BJAB cells. BJAB cells mock transfected (a) or transfected with 10 μg of pEGFP-C1 EBNA2 (b, c, and d) were immunostained by using anti-SMN 7B10/anti-mouse TRITC MAbs and subjected to confocal laser scanning microscopy. In the merged image (subpanel d), colocalization results in a yellow signal.
FIG. 3
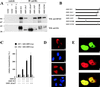
Enhanced coactivation of the LMP1 promoter by DP103 binding-deficient SMN ΔEx6/7. (A) Mapping of the DP103 binding site on SMN. 293GP cells were transfected with pSG5 constructs encoding HA-tagged SMN mutants (10 μg) as indicated. After 36 h native cell extracts were immunoprecipitated with anti-HA MAb 3F10 (IP: anti HA and control 2) or unspecific MAb 3A6 (controls 1 and 3) and analyzed by SDS–10% PAGE and Western blotting. Precipitated transfected SMN mutants were detected by using anti-HA MAb 3F10 (WB: anti HA), and coprecipitated endogenous DP103 was detected by using anti-DP103 MAb 8H4 (WB: anti DP103). The positions of the molecular mass markers (in kilodaltons) are indicated on the left side of each panel. Deletion of SMN exon 6 abolished coprecipitation of endogenous DP103. (B) Schematic representation of the SMN mutants tested. (C) Coexpression of EBNA2 and the HA-tagged DP103 binding-deficient SMN mutant SMN ΔEx6/7 further increased coactivation of the −327/+40 LMP1 promoter luciferase construct. Assays were performed as described for Fig. 1B. Graphs represent the mean values of three independent experiments performed in duplicate (±SEM). (D) Immunofluorescence of HA-tagged WT SMN (a) and SMN ΔEx6/7 (c) expressed in HeLa cells and stained with 3F10 anti-HA/anti-rat TRITC MAbs. (b and d) Nuclei were visualized by using DAPI (4′,6′-diamidino-2-phenylindole). Loss of binding to DP103 released SMN ΔEx6/7 from nuclear gems/coiled bodies. (E) Enhanced colocalization of EBNA2 and DP103 binding-deficient SMN ΔEx6/7. HeLa cells coexpressing EGFP-EBNA2 (a) and HA-tagged SMN ΔEx6/7 (b) were stained by using anti-HA 3F10/anti-rat TRITC MAbs and subjected to confocal laser scanning microscopy. In the merged image (subpanel c), colocalization results in a yellow signal.
FIG. 4
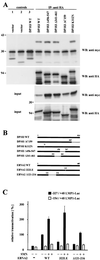
EBNA2-mediated transactivation of the LMP1 promoter involves binding of EBNA2 to DP103. (A) Mapping of the SMN binding site on DP103. 293GP cells were transfected with pSG5 constructs encoding HA-tagged DP103 mutants and WT myc-tagged SMN, as indicated. Cell extracts were immunoprecipitated with anti-HA MAb 3F10 (IP: anti HA and control 2) or irrelevant MAb 9C2 (controls 1 and 3) and analyzed by SDS–10% PAGE and Western blotting. Precipitated HA-tagged DP103 mutants were detected by using anti-HA MAb 3F10 (WB: anti HA), coprecipitated myc-tagged WT SMN by using anti-myc MAb 9E10 (WB: anti myc). Panels designated input represent ca. 10% of unprecipitated extracts. The positions of the molecular mass markers (in kilodaltons) are indicated on the left side of each panel. Deletion of aa 665 to 824 of DP103 (ΔC159) abolished coprecipitation of myc-tagged SMN. (B) Schematic representation of the DP103 and EBNA2 mutants tested. (C) Coexpression of SMN rescued impaired transactivation of the −327/+40 LMP1 promoter luciferase construct by the DP103 binding-deficient mutant EBNA2 Δ121-216 and the RBPJκ binding-deficient mutant EBNA2 322LE. Assays were performed as described for Fig. 1B. Graphs represent the mean values of three independent experiments performed in duplicate (±SEM).
FIG. 5
The presence of RBPJκ is essential but not sufficient for coactivation by EBNA2 and SMN. (A) Schematic map of positions −327 to +40 of the LMP1 promoter and the different luciferase constructs tested. Shaded boxes represent the positions of cellular transcription factor binding sites, and black boxes represent the positions RBPJκ binding sites relative to the transcription start site (arrow). (B) Deletion mutants of the LMP1 promoter were tested for responsiveness to EBNA2 and coexpressed HA-tagged SMN in BJAB cells, as indicated. Deletion of the RBPJκ binding sites abolished EBNA2 transactivation and coactivation by SMN. (C) No coactivation of a multimerized RBPJκ site by SMN and increasing amounts of WT EBNA2 (1, 4, and 10 μg) or by SMN and RBPJκ binding-deficient EBNA2 322LE. (B and C) Assays were performed as described for Fig. 1B. Graphs represent the mean values of three independent experiments performed in duplicate (±SEM).
Similar articles
- Epstein-Barr virus nuclear antigen 2-estrogen receptor fusion proteins transactivate viral and cellular genes and interact with RBP-J kappa in a conditional fashion.
Kempkes B, Pawlita M, Zimber-Strobl U, Eissner G, Laux G, Bornkamm GW. Kempkes B, et al. Virology. 1995 Dec 20;214(2):675-9. doi: 10.1006/viro.1995.0084. Virology. 1995. PMID: 8553575 - Domains of the Epstein-Barr virus nuclear antigen 2 (EBNA2) involved in the transactivation of the latent membrane protein 1 and the EBNA Cp promoters.
Sjöblom A, Nerstedt A, Jansson A, Rymo L. Sjöblom A, et al. J Gen Virol. 1995 Nov;76 ( Pt 11):2669-78. doi: 10.1099/0022-1317-76-11-2669. J Gen Virol. 1995. PMID: 7595374 - The SMN complex.
Gubitz AK, Feng W, Dreyfuss G. Gubitz AK, et al. Exp Cell Res. 2004 May 15;296(1):51-6. doi: 10.1016/j.yexcr.2004.03.022. Exp Cell Res. 2004. PMID: 15120993 Review. - EBNA2 and Notch signalling in Epstein-Barr virus mediated immortalization of B lymphocytes.
Zimber-Strobl U, Strobl LJ. Zimber-Strobl U, et al. Semin Cancer Biol. 2001 Dec;11(6):423-34. doi: 10.1006/scbi.2001.0409. Semin Cancer Biol. 2001. PMID: 11669604 Review.
Cited by
- The nuclear chaperone nucleophosmin escorts an Epstein-Barr Virus nuclear antigen to establish transcriptional cascades for latent infection in human B cells.
Liu CD, Chen YL, Min YL, Zhao B, Cheng CP, Kang MS, Chiu SJ, Kieff E, Peng CW. Liu CD, et al. PLoS Pathog. 2012;8(12):e1003084. doi: 10.1371/journal.ppat.1003084. Epub 2012 Dec 13. PLoS Pathog. 2012. PMID: 23271972 Free PMC article. - Interferon regulatory factor 7 regulates expression of Epstein-Barr virus latent membrane protein 1: a regulatory circuit.
Ning S, Hahn AM, Huye LE, Pagano JS. Ning S, et al. J Virol. 2003 Sep;77(17):9359-68. doi: 10.1128/jvi.77.17.9359-9368.2003. J Virol. 2003. PMID: 12915551 Free PMC article. - Global Phosphoproteomics Analysis of IBRS-2 Cells Infected With Senecavirus A.
Li J, Zhang Z, Lv J, Ma Z, Pan L, Zhang Y. Li J, et al. Front Microbiol. 2022 Jan 26;13:832275. doi: 10.3389/fmicb.2022.832275. eCollection 2022. Front Microbiol. 2022. PMID: 35154063 Free PMC article. - Use of antibodies against Epstein-Barr virus nuclear antigen 1 for detection of cellular proteins with monomethylated arginine residues that are potentially involved in viral transformation.
Graesser C, Nord R, Flaswinkel H, Kremmer E, Meese E, Caban KM, Fröhlich T, Grässer FA, Hart M. Graesser C, et al. Arch Virol. 2024 Nov 8;169(12):241. doi: 10.1007/s00705-024-06172-7. Arch Virol. 2024. PMID: 39514105 Free PMC article. - Interferon regulatory factor 5 represses expression of the Epstein-Barr virus oncoprotein LMP1: braking of the IRF7/LMP1 regulatory circuit.
Ning S, Huye LE, Pagano JS. Ning S, et al. J Virol. 2005 Sep;79(18):11671-6. doi: 10.1128/JVI.79.18.11671-11676.2005. J Virol. 2005. PMID: 16140744 Free PMC article.
References
- Campbell L, Hunter C M D, Mohagheg P, Tinsley J M, Brasch M A, Davies K E. Direct interaction of SMN with dp103, a putative RNA helicase: a role for SMN in transcription regulation? Hum Mol Genet. 2000;9:1093–1100. - PubMed
- Clermont O, Burlet P, Cruad C, Bertrandy S, Melki J, Munnich A, Lefevbre S. Mutation analysis of the SMN gene in undeleted SMA patients. Am J Hum Genet. 1997;61:A329.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources