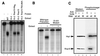Rna15 interaction with the A-rich yeast polyadenylation signal is an essential step in mRNA 3'-end formation - PubMed (original) (raw)
Rna15 interaction with the A-rich yeast polyadenylation signal is an essential step in mRNA 3'-end formation
S Gross et al. Mol Cell Biol. 2001 Dec.
Abstract
In Saccharomyces cerevisiae, four factors [cleavage factor I (CF I), CF II, polyadenylation factor I (PF I), and poly(A) polymerase (PAP)] are required for maturation of the 3' end of the mRNA. CF I and CF II are required for cleavage; a complex of PAP and PF I, which includes CF II subunits, participates in polyadenylation, along with CF I. These factors are directed to the appropriate site on the mRNA by two sequences: one A-rich and one UA-rich. CF I contains five proteins, two of which, Rna15 and Hrp1, interact with the mRNA through RNA recognition motif-type RNA binding motifs. Previous work demonstrated that the UV cross-linking of purified Hrp1 to RNA required the UA-rich element, but the contact point of Rna15 was not known. We show here that Rna15 does not recognize a particular sequence in the absence of other proteins. However, in complex with Hrp1 and Rna14, Rna15 specifically interacts with the A-rich element. The Pcf11 and Clp1 subunits of CF I are not needed to position Rna15 at this site. This interaction is essential to the function of CF I. A mutant Rna15 with decreased affinity for RNA is defective for in vitro RNA processing and lethal in vivo, while an RNA with a mutation in the A-rich element is not processed in vitro and can no longer be UV cross-linked to the Rna15 subunit assembled into CF I. Thus, the recognition of the A-rich element depends on the tethering of Rna15 through an Rna14 bridge to Hrp1 bound to the UA-rich motif. These results illustrate that the yeast 3' end is defined and processed by a mechanism surprisingly different from that used by the mammalian system.
Figures
FIG. 1
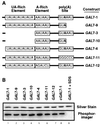
Cross-linking of CF I proteins to GAL7 3′-UTR in vitro demonstrates the binding of Rna15 and Hrp1. Subcomplexes or complete CF I were assembled from purified proteins as described in Materials and Methods and cross-linked to the GAL7-1 RNA precursor by using UV irradiation. The presence of a plus sign indicates the addition of that protein to the assembly reaction. (A) After cross-linking and digestion of the RNA, proteins were resolved by SDS-PAGE and visualized by silver staining, and radioactively tagged proteins were identified by PhosphorImager analysis. (B) Determination of the identity of the 70-kDa radioactively tagged protein. After photo-cross-linking, complexes containing both Hrp1 and Rna14 were disrupted, and proteins were immunoprecipitated by using either anti-Hrp1 or anti-Rna14 antisera, indicated by the Hrp1 or Rna14 labels. Captured proteins were released by boiling in SDS buffer and identified by Western blotting by using antibodies to the His6 epitope found on both proteins.
FIG. 2
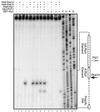
Rna15 alone does not recognize a specific RNA sequence in the absence of other CF I proteins. (A) Schematic of GAL7 3′-UTR and mutant derivatives utilized in the cross-linking study. RNA is indicated by a solid line, deletions are indicated by the absence of the line, conserved sequences are boxed, and the cleavage site is indicated by the boldface adenosine residue immediately upstream. (B) Silver stain and phosphorimager scan from gel containing purified Rna15 after UV cross-linking to the RNAs indicated. cGAL7-1, transcript derived from the complementary strand to the GAL7 3′-UTR; GAL7-1 + SDS, cross-linking reaction was done under denaturing conditions.
FIG. 3
Mapping of the binding sites of Hrp1 and Rna15. Subcomplexes or complete CF I was assembled from purified active proteins and photo-cross-linked to GAL7-1 RNA; a plus sign indicates the addition of a particular protein to the assembly reaction. Cross-linking of Hrp1 to the UA-rich element creates a polymerase pause site at the 14th nucleotide of the UA-rich element (open arrow). Specific binding of Rna15 in complex with Hrp1 and Rna14 creates a polymerase pause site at the sixth nucleotide of the A-rich element (closed arrow).
FIG. 4
The Rna15 F63,66A mutant does not disrupt assembly of the CF IA complex. (A) Clarified whole-cell lysates of insect cells either mock infected (Mock) or infected with recombinant baculoviruses expressing either wild-type or the F63,66A mutant Rna15 proteins (indicated by the arrow). The protein immediately below the Rna15 is a 32-kDa baculovirus structural protein. (B) GST-Pcf11 bound to glutathione-Sepharose beads was used to nucleate the assembly of CF IA incorporating either Rna15-His6 purified from E. coli or insect cell lysates from cells expressing wild-type or mutant Rna15. The plus symbol indicates the presence of a particular protein in the assembly reaction. Proteins bound to the beads through interaction with GST-Pcf11 were released by the addition of glutathione and visualized by Western blotting with the indicated antisera. The “10% input” lane represents direct loading on the gel of a quantity of protein equal to 10% of the amount added to the complex assembly reaction.
FIG. 5
The Rna15 F63,66A mutant displays reduced affinity for RNA, as demonstrated by a poly(U) binding assay with wild-type and mutant Rna15 proteins. Proteins in the lysates from Fig. 4A were assayed for the ability to bind to poly(U) Sepharose beads in the presence of increasing concentrations of KCl and then detected by Western blotting with Rna15 antibody.
FIG. 6
Disruption of the interaction between the A-rich element and Rna15 prevents RNA 3′-end processing in vitro. (A) Rna15 F63,66A cannot rescue the processing activity of rna15-2 mutant extract. Full-length radioactive GAL7-1 RNA was incubated with extract made from wild-type cells (WT) or from cells carrying the rna15-2 mutation (15-2) under standard processing conditions. As indicated, reactions were supplemented with Rna15-His6 protein purified from E. coli or with lysates of insect cells expressing wild-type or mutant Rna15. RNAs recovered from the reactions were separated by electrophoresis through a 5% acrylamide-bisacrylamide (19:1) gel supplemented with 8 M urea. Lane 1 shows unreacted precursor. (B) The A-rich element is essential for 3′-end processing of RNA in vitro. The addition of yeast whole-cell extract (indicated by a plus sign) results in the cleavage and polyadenylation of the GAL7-1 precursor. When this sequence is mutated from AAUAAU to AGAUCU in GAL7-12, this processing is abolished. (C) Mutation of the A-rich element prevents Rna15 binding to the RNA. Proteins in a Q-Sepharose fraction containing CF I activity were photo-cross-linked to either wild-type (wt) RNA or RNA containing a mutation in the A-rich sequence (M). After digestion of the RNA, samples were immunoprecipitated by using combined anti-Rna15 and anti-Hrp1 antiserum or a control antibody (C) as indicated. Proteins were resolved by SDS-PAGE, transferred to membrane and then detected by Western blotting or autoradiography.
FIG. 7
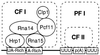
Model for interaction of the polyadenylation complex with conserved sequence elements in the pre-mRNA. The CF I architecture is taken from the protein-protein interaction studies of Gross and Moore (23).
Similar articles
- Five subunits are required for reconstitution of the cleavage and polyadenylation activities of Saccharomyces cerevisiae cleavage factor I.
Gross S, Moore C. Gross S, et al. Proc Natl Acad Sci U S A. 2001 May 22;98(11):6080-5. doi: 10.1073/pnas.101046598. Epub 2001 May 8. Proc Natl Acad Sci U S A. 2001. PMID: 11344258 Free PMC article. - Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3'-end formation in yeast.
Kessler MM, Henry MF, Shen E, Zhao J, Gross S, Silver PA, Moore CL. Kessler MM, et al. Genes Dev. 1997 Oct 1;11(19):2545-56. doi: 10.1101/gad.11.19.2545. Genes Dev. 1997. PMID: 9334319 Free PMC article. - Architectural and functional details of CF IA proteins involved in yeast 3'-end pre-mRNA processing and its significance for eukaryotes: A concise review.
Kaur M, Sharma A, Singh G, Kumar S, Barnwal RP. Kaur M, et al. Int J Biol Macromol. 2021 Dec 15;193(Pt A):387-400. doi: 10.1016/j.ijbiomac.2021.10.129. Epub 2021 Oct 23. Int J Biol Macromol. 2021. PMID: 34699898 Review. - Implications of polyadenylation in health and disease.
Curinha A, Oliveira Braz S, Pereira-Castro I, Cruz A, Moreira A. Curinha A, et al. Nucleus. 2014;5(6):508-19. doi: 10.4161/nucl.36360. Epub 2014 Oct 31. Nucleus. 2014. PMID: 25484187 Free PMC article. Review.
Cited by
- RNA polymerase II CTD phosphopeptides compete with RNA for the interaction with Pcf11.
Hollingworth D, Noble CG, Taylor IA, Ramos A. Hollingworth D, et al. RNA. 2006 Apr;12(4):555-60. doi: 10.1261/rna.2304506. Epub 2006 Feb 22. RNA. 2006. PMID: 16497660 Free PMC article. - Yeast Sen1 helicase protects the genome from transcription-associated instability.
Mischo HE, Gómez-González B, Grzechnik P, Rondón AG, Wei W, Steinmetz L, Aguilera A, Proudfoot NJ. Mischo HE, et al. Mol Cell. 2011 Jan 7;41(1):21-32. doi: 10.1016/j.molcel.2010.12.007. Mol Cell. 2011. PMID: 21211720 Free PMC article. - Unphosphorylated SR-like protein Npl3 stimulates RNA polymerase II elongation.
Dermody JL, Dreyfuss JM, Villén J, Ogundipe B, Gygi SP, Park PJ, Ponticelli AS, Moore CL, Buratowski S, Bucheli ME. Dermody JL, et al. PLoS One. 2008 Sep 26;3(9):e3273. doi: 10.1371/journal.pone.0003273. PLoS One. 2008. PMID: 18818768 Free PMC article. - How is polyadenylation restricted to 3'-untranslated regions?
Struhl K. Struhl K. Yeast. 2024 Apr;41(4):186-191. doi: 10.1002/yea.3915. Epub 2023 Dec 2. Yeast. 2024. PMID: 38041485 Review. - A genome-wide screen for essential yeast genes that affect telomere length maintenance.
Ungar L, Yosef N, Sela Y, Sharan R, Ruppin E, Kupiec M. Ungar L, et al. Nucleic Acids Res. 2009 Jul;37(12):3840-9. doi: 10.1093/nar/gkp259. Epub 2009 Apr 22. Nucleic Acids Res. 2009. PMID: 19386622 Free PMC article.
References
- Bandziulis R J, Swanson M S, Dreyfuss G. RNA-binding proteins as developmental regulators. Genes Dev. 1989;3:431–437. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials