Pattern and timing of gene duplication in animal genomes - PubMed (original) (raw)
Comparative Study
. 2001 Nov;11(11):1842-7.
doi: 10.1101/gr.200601.
Affiliations
- PMID: 11691848
- PMCID: PMC311158
- DOI: 10.1101/gr.200601
Comparative Study
Pattern and timing of gene duplication in animal genomes
R Friedman et al. Genome Res. 2001 Nov.
Abstract
Duplication of genes, giving rise to multigene families, has been a characteristic feature of the evolution of eukaryotic genomes. In the case of vertebrates, it has been proposed that an increase in gene number resulted from two rounds of duplication of the entire genome by polyploidization (the 2R hypothesis). In the most extensive test to date of this hypothesis, we compared gene numbers in homologous families and conducted phylogenetic analyses of gene families with two to eight members in the complete genomes of Caenorhabditis elegans and Drosophila melanogaster and the available portion of the human genome. Although the human genome showed a higher proportion of recent gene duplications than the other animal genomes, the proportion of duplications after the deuterostome-protostome split was constant across families, with no peak of such duplications in four-member families, contrary to the expectation of the 2R hypothesis. A substantial majority (70.9%) of human four-member families and four-member clusters in larger families showed topologies inconsistent with two rounds of polyploidization in vertebrates.
Figures
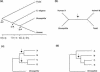
Figure 1
(a) Phylogenetic tree showing the major cladogenetic events used in timing gene duplications: (A–F) the animal–fungus divergence; (C–N), the coelomate–nematode divergence; (D–P), the deuterostome–protostome divergence. The topology of the tree and the divergence time estimates (±SE) are from Wang et al. (1999). However, gene duplications were timed relative to cladogenetic events independent of a molecular clock assumption. (b) Hypothetical gene family containing two human members (A and B). If the internal branch (indicated by arrow) is significantly supported, we can conclude (independent of the rooting of the tree) that A and B diverged prior to the deuterostome–protostome divergence. (c) Hypothetical four-member human gene family having a topology of the form (AB) (CD) consistent with the hypothesis of two rounds of genome duplication (the 2R hypothesis). (d) Hypothetical four-member human gene family having a topology of the form (A) (BD) inconsistent with the 2R hypothesis.
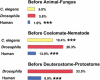
Figure 2
Numbers of gene duplications in two- to eight-member families. A duplication was dated prior to one of the three a major cladogenetic events (the animal–fungus divergence, the coelomate–nematode divergence, and the deuterostome–protostome divergence) if its occurrence prior to the event was supported by a significant internal branch. Chi square tests of the hypothesis that the proportion of duplications prior to a cladogenetic event differed from that in Drosophila: (***) P < 0.001. Numbers of duplication events were as follows: Caenorhabditis elegans, 463; Drosophila 567; human, 1760.
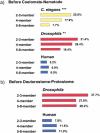
Figure 3
Numbers of gene duplications in three family size classes (two- to three-member families, four-member families, five- to eight-member families). A duplication was dated prior to one of two major cladogenetic events, (a) the coelomate–nematode divergence, and (b) the deuterostome–protostome divergence, if its occurrence prior to the event was supported by a significant internal branch. Chi-square tests of the uniformity across family size classes of the proportion of duplications prior to the cladogenetic event : (**) P < 0.01; (***) P< 0.001.
Similar articles
- New evidence for genome-wide duplications at the origin of vertebrates using an amphioxus gene set and completed animal genomes.
Panopoulou G, Hennig S, Groth D, Krause A, Poustka AJ, Herwig R, Vingron M, Lehrach H. Panopoulou G, et al. Genome Res. 2003 Jun;13(6A):1056-66. doi: 10.1101/gr.874803. Genome Res. 2003. PMID: 12799346 Free PMC article. - Nematode and arthropod genomes provide new insights into the evolution of class 2 B1 GPCRs.
Cardoso JC, Félix RC, Power DM. Cardoso JC, et al. PLoS One. 2014 Mar 20;9(3):e92220. doi: 10.1371/journal.pone.0092220. eCollection 2014. PLoS One. 2014. PMID: 24651821 Free PMC article. - Gene duplication and the structure of eukaryotic genomes.
Friedman R, Hughes AL. Friedman R, et al. Genome Res. 2001 Mar;11(3):373-81. doi: 10.1101/gr.155801. Genome Res. 2001. PMID: 11230161 Free PMC article. - Detection of gene duplications and block duplications in eukaryotic genomes.
Li WH, Gu Z, Cavalcanti AR, Nekrutenko A. Li WH, et al. J Struct Funct Genomics. 2003;3(1-4):27-34. J Struct Funct Genomics. 2003. PMID: 12836682 Review. - Genome duplication and gene-family evolution: the case of three OXPHOS gene families.
De Grassi A, Lanave C, Saccone C. De Grassi A, et al. Gene. 2008 Sep 15;421(1-2):1-6. doi: 10.1016/j.gene.2008.05.011. Epub 2008 Jun 23. Gene. 2008. PMID: 18573316 Review.
Cited by
- Selectionism and neutralism in molecular evolution.
Nei M. Nei M. Mol Biol Evol. 2005 Dec;22(12):2318-42. doi: 10.1093/molbev/msi242. Epub 2005 Aug 24. Mol Biol Evol. 2005. PMID: 16120807 Free PMC article. - Genome duplication, a trait shared by 22000 species of ray-finned fish.
Taylor JS, Braasch I, Frickey T, Meyer A, Van de Peer Y. Taylor JS, et al. Genome Res. 2003 Mar;13(3):382-90. doi: 10.1101/gr.640303. Genome Res. 2003. PMID: 12618368 Free PMC article. - New evidence for genome-wide duplications at the origin of vertebrates using an amphioxus gene set and completed animal genomes.
Panopoulou G, Hennig S, Groth D, Krause A, Poustka AJ, Herwig R, Vingron M, Lehrach H. Panopoulou G, et al. Genome Res. 2003 Jun;13(6A):1056-66. doi: 10.1101/gr.874803. Genome Res. 2003. PMID: 12799346 Free PMC article. - A genetic linkage map and comparative genome analysis of common carp (Cyprinus carpio L.) using microsatellites and SNPs.
Zheng X, Kuang Y, Zhang X, Lu C, Cao D, Li C, Sun X. Zheng X, et al. Mol Genet Genomics. 2011 Oct;286(3-4):261-77. doi: 10.1007/s00438-011-0644-x. Epub 2011 Aug 26. Mol Genet Genomics. 2011. PMID: 21870156 - Evolution of glucose utilization: glucokinase and glucokinase regulator protein.
Irwin DM, Tan H. Irwin DM, et al. Mol Phylogenet Evol. 2014 Jan;70:195-203. doi: 10.1016/j.ympev.2013.09.016. Epub 2013 Sep 25. Mol Phylogenet Evol. 2014. PMID: 24075984 Free PMC article. Review.
References
- Bork RP, Copley R. Filling in the gaps. Nature. 2001;409:818–820. - PubMed
- Guigo R, Muchnik I, Smith TF. Reconstruction of ancient molecular phylogeny. Mol Phyl Evol. 1996;46:189–213. - PubMed
- Hughes AL. Phylogenetic tests of the hypothesis of block duplication of homologous genes on human chromosomes 6, 9, and 1. Mol Biol Evol. 1998;15:854–870. - PubMed
- ————— . Adaptive evolution of genes and genomes. New York: Oxford University Press; 1999a.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases