Cell cycle-dependent degradation of the Saccharomyces cerevisiae spindle motor Cin8p requires APC(Cdh1) and a bipartite destruction sequence - PubMed (original) (raw)
Cell cycle-dependent degradation of the Saccharomyces cerevisiae spindle motor Cin8p requires APC(Cdh1) and a bipartite destruction sequence
E R Hildebrandt et al. Mol Biol Cell. 2001 Nov.
Free PMC article
Abstract
Saccharomyces cerevisiae Cin8p belongs to the BimC family of kinesin-related motor proteins that are essential for spindle assembly. Cin8p levels were found to oscillate in the cell cycle due in part to a high rate of degradation imposed from the end of mitosis through the G1 phase. Cin8p degradation required the anaphase-promoting complex ubiquitin ligase and its late mitosis regulator Cdh1p but not the early mitosis regulator Cdc20p. Cin8p lacks a functional destruction box sequence that is found in the majority of anaphase-promoting complex substrates. We carried out an extensive mutagenesis study to define the cis-acting sequence required for Cin8p degradation in vivo. The C terminus of Cin8p contains two elements required for its degradation: 1) a bipartite destruction sequence composed of a KEN-box plus essential residues within the downstream 22 amino acids and 2) a nuclear localization signal. The bipartite destruction sequence appears in other BimC kinesins as well. Expression of nondegradable Cin8p showed very mild phenotypic effects, with an increase in the fraction of mitotic cells with broken spindles.
Figures
Figure 1
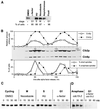
Cin8p oscillates and is unstable in G1. (A) Cin8p is absent from G1 cells. Yeast cells (MAY4823) expressing genomic CIN8-3HA were treated with α-factor, hydroxyurea (HU), nocodazole, and DMSO only for 3.7 h. Asynchronous cells were applied to a Ficoll gradient for collection of small G1 cells. The percentage of cells with the morphology characteristic of the respective cell cycle arrest is shown below the blot. (B) Cin8p levels fluctuate in the cell cycle. Yeast cells expressing genomic CIN8-3HA (MAY4822) were synchronized with α-factor in G1. Cells were washed and released into medium without α-factor. Samples were collected every 10 min and analyzed for Cin8p and Clb2p levels by Western blot (middle) and for the presence of bipolar spindles by tubulin immunofluorescence (bottom). The top panel shows quantification of Cin8p and Clb2p levels. (C) Cin8p is unstable in G1. Strain K1534 carrying a P_GAL1_ > CIN8-3HA plasmid was grown in medium with 2% raffinose. Cells were treated with α-factor, hydroxyurea, nocodazole, or DMSO for 3 h. Cin8p was then induced by incubation with 2% galactose, followed by addition of cycloheximide and glucose (t = 0 minutes). Samples were collected at the indicated times (minutes), and protein extracts were prepared. Equal amounts of protein were separated by gel electrophoresis and analyzed for Cin8p levels by Western blotting. (D) Cin8p is stable in late anaphase. A cdc15-2 (MAY4751) strain carrying a P_GAL1_ > 6myc-SV40NLS-CIN8 plasmid was grown in synthetic medium plus raffinose at 22°C. Part of the culture was shifted to 37°C for 4.5 h to arrest in late anaphase. Another portion was treated with α-factor at 22°C for 4 h and then shifted to 37°C for 30 min to inactivate Cdc15-2p. Cin8p was induced with galactose for 90 min, followed by addition of cycloheximide and glucose (t = 0). In the α-factor–treated sample nearly all of the Cin8p was degraded by the time the first time sample was processed.
Figure 2
Cin8p degradation requires the APC subunit Cdc16p and the regulator Cdh1p but not Cdc20p. (A) Cin8p's instability in G1 requires CDC16, not CDC20. Wild-type (K1534, WT), cdc16-123 (K4438), and cdc20-1 (MAY5402) strains with P_GAL1_ > CIN8-3HA were examined by the G1 expression-pulse protocol described in Figure 1C except that the temperature was increased to 37°C beginning 30 min before the Cin8p pulse. (B) Cin8p is stabilized in G1-arrested cdh1Δ cells. Wild-type (K1534), cdh1Δ (MAY5353), clb2Δ (MAY6810), and cdh1Δ clb2Δ (MAY6812) strains with P_GAL1_ > CIN8-3HA were examined by the G1 expression-pulse protocol described in Figure 1C. (C) Cin8p is absent from G1 cells isolated from an asynchronous culture. Unbudded wild-type (MAY4822) and cdh1Δ (MAY5466) cells with genomic CIN8-3HA were isolated from a Ficoll gradient and analyzed for Cin8p and Clb2p levels. The asterisk indicates a background band recognized by the Clb2 antibody. (D) Moderate overexpression of CIN8 kills cdh1Δ but not cdc20-1 cells. Wild-type (MAY4822), cdh1Δ (MAY5466), and cdc20-1 (MAY5017) with P_GALS_ > CIN8 were spotted in serial dilution (left to right) onto solid synthetic medium containing either glucose (CIN8 expression off) or galactose (CIN8 expression on) and grown at 26°C.
Figure 3
Cin8p degradation is
d
-box independent but requires localization to the nucleus. (A) Schematic of Cin8p domains drawn to scale. The potential
d
-box and KEN-box sequences are indicated as well as the region required for Cin8p nuclear localization. (B) Deletion of the
d
-box does not stabilize Cin8p. Wild-type (K1534) cells with plasmids expressing full-length Cin8p (P_GAL1_ > CIN8-3HA) or Cin8p with the
d
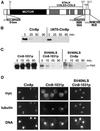
-box deleted (P_GAL1_ > ΔN70-CIN8-3HA) were subjected to the G1 expression-pulse protocol described in Figure 1C. (C) Cin8p degradation requires a functional NLS. A wild-type strain (K1534) carrying plasmids expressing Cin8p derivatives with Cin8p's C-terminal NLS and/or an N-terminal SV40NLS (P_GAL1_ > 6myc-CIN8-1031; P_GAL1_ > 6myc-SV40NLS-CIN8-1031; P_GAL1_ > 6myc-SV40NLS-CIN8, from left to right) were subjected to the G1 expression-pulse protocol described in Figure 1C. (D) Demonstration that the SV40NLS restores nuclear localization to CIN8-1031. A cin8Δ kip1Δ strain (MAY5590) carrying 6myc-tagged versions of CIN8 on high-copy plasmids (6myc-CIN8; 6myc-CIN8-1031; 6myc-SV40NLS-CIN8-1031, from left to right) were arrested at the short spindle stage with hydroxyurea and stained for Cin8p (myc), microtubules (tubulin), and DNA. Although most of Cin8-1031p is cytoplasmic, a portion still binds spindles.
Figure 4
Cin8p's degradation signal is in the C terminus. Wild-type (K1534, WT) cells expressing versions of P_GAL1_ > 6myc-SV40NLS-CIN8, truncated at the indicated amino acid, were subjected to the G1 expression-pulse protocol described in Figure 1C.
Figure 5
Moderate overexpression and stability of random and site-directed Cin8p mutants. Select random (A) and site-directed (B) Cin8p mutants were tested for toxicity when moderately overexpressed from P_GALS_ (P_GALS_ assay) and for Cin8p stability in vivo (degradation assay). P_GALS_ assay: Wild-type (MAY2422, WT) cells carrying mutant derivatives of P_GALS_ > CIN8 were spotted onto solid synthetic medium with either glucose or galactose (as in Figure 2D except a single dilution spot is shown). Degradation assay: P_GAL1_ > 6myc-SV40NLS-CIN8 derivatives of the mutants were tested for stability in strain K1534 by the G1 expression-pulse protocol described in Figure 1C. The figures on the right depict the positions of mutant sites in the amino acid 956-1015 region of Cin8p. The boundaries of element 2 are defined in Figure 8A. Precise sequence changes are described in Figure 6. (C) Wild-type (MAY2422) cells carrying mutant derivatives of P_GALS_ > CIN8 were spotted (as in Figure 2D except two dilution spots are shown) onto solid synthetic medium with either glucose or galactose plus 0.05% glucose as carbon sources.
Figure 6
Summary of mutants in the region of KEN-box and element 2. Summary of findings from the two assays (P_GALS_ and degradation assay) to test the stability of random and site-directed Cin8p mutants. A “+” under P_GALS_ indicates wild-type levels of growth on galactose-containing media, and a “−” indicates death on galactose. A “+” in the degradation column (Degr.) indicates a wild-type (WT) level of instability, and a “−” indicates that the mutant was stable. “n.d.” means the mutant was not tested. Numbers given in parentheses (e.g., x2) in the Cin8p column indicate the number of times a particular random mutant was isolated. Amino acid positions are indicated at the top of the right column. The KEN-box and element 2 are boxed.
Figure 7
The C terminus of Cin8p destabilizes a heterologous protein. (A) Schematic of Gal4BD-Cin8p fusion (to scale). (B) The C terminus of Cin8p is necessary and sufficient for degradation. Wild-type (K1534, WT) or cdh1Δ (MAY5353) strains carrying plasmids expressing Gal4BD (P_GAL1_ > GAL4BD) or Gal4BD fused to a C-terminal portion of wild-type Cin8p (P_GAL1_ > GAL4BD-cin8), Cin8-Δ996-1004p (P_GAL1_ > GAL4BD-cin8-Δ996-1004), or Cin8-alaKENp (P_GAL1_ > GAL4BD-cin8-alaKEN) were subjected to the to the G1 expression-pulse protocol described in Figure 1C. Samples were collected 1–5 min before the end of the galactose pulse (−1 or −5 min) and at times indicated after the pulse. The wild-type fusion was nearly completely degraded by the time the first time sample was processed. The positions of molecular weight standards are shown on the left. The Western blot was probed with anti-Gal4BD antibody.
Figure 8
The destruction sequence in Cin8p and possibly other BimC motors is bipartite. (A) Summary of specific amino acid contributions to Cin8p's degradation signal. The C-terminal side of element 2, amino acid 998, is defined by the shortest truncated form that still supports degradation. The N-terminal side is defined by the positions of the random non–KEN-box mutants (Figure 6B). Note that the borders of element 2 are arbitrary because surrounding amino acids are also important. “Essential” amino acids are those with a single point mutant-stabilized Cin8p. “Contributes” to degradation means the highlighted amino acid had a partial effect when individually mutated or Cin8p was completely stabilized when the residue was mutated in combination with other amino acids. Underlined residues show a region that partially affects Cin8p degradation when deleted or scrambled. (B) Alignment of BimC motors that have a KEN-box–like sequence in the C-terminal domain, after the last coiled-coil. The alignment was performed on sequences starting at the KEN-box (amino acid positions indicated) with the use of the PIMA 1.4 (Smith and Smith, 1992) and BoxShade 3.21 alignment programs and final manual adjustment. Asterisks represent the C terminus of the protein. In the consensus line, capital letters represent identical residues, and lower case letters represent conserved residues. Greater than 40% of sequences must agree for shading. Sc, S. cerevisiae; Xl, Xenopus laevis; Hs, Homo sapiens; Mm, Mus musculus; Pl, Paracentrotus lividus; Spu, Strongylocentrotus purpuratus; At, Arabidopsis thaliana; Nt, Nicotiana tabacum; Dm, Drosophila melanogaster.
Similar articles
- D box and KEN box motifs in budding yeast Hsl1p are required for APC-mediated degradation and direct binding to Cdc20p and Cdh1p.
Burton JL, Solomon MJ. Burton JL, et al. Genes Dev. 2001 Sep 15;15(18):2381-95. doi: 10.1101/gad.917901. Genes Dev. 2001. PMID: 11562348 Free PMC article. - Degradation of the kinesin Kip1p at anaphase onset is mediated by the anaphase-promoting complex and Cdc20p.
Gordon DM, Roof DM. Gordon DM, et al. Proc Natl Acad Sci U S A. 2001 Oct 23;98(22):12515-20. doi: 10.1073/pnas.231212498. Epub 2001 Oct 16. Proc Natl Acad Sci U S A. 2001. PMID: 11606759 Free PMC article. - The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1.
Pfleger CM, Kirschner MW. Pfleger CM, et al. Genes Dev. 2000 Mar 15;14(6):655-65. Genes Dev. 2000. PMID: 10733526 Free PMC article. - Mitotic motors in Saccharomyces cerevisiae.
Hildebrandt ER, Hoyt MA. Hildebrandt ER, et al. Biochim Biophys Acta. 2000 Mar 17;1496(1):99-116. doi: 10.1016/s0167-4889(00)00012-4. Biochim Biophys Acta. 2000. PMID: 10722880 Review. - Control of mitotic transitions by the anaphase-promoting complex.
Fang G, Yu H, Kirschner MW. Fang G, et al. Philos Trans R Soc Lond B Biol Sci. 1999 Sep 29;354(1389):1583-90. doi: 10.1098/rstb.1999.0502. Philos Trans R Soc Lond B Biol Sci. 1999. PMID: 10582244 Free PMC article. Review.
Cited by
- Building a Regulatory Network with Short Linear Sequence Motifs: Lessons from the Degrons of the Anaphase-Promoting Complex.
Davey NE, Morgan DO. Davey NE, et al. Mol Cell. 2016 Oct 6;64(1):12-23. doi: 10.1016/j.molcel.2016.09.006. Mol Cell. 2016. PMID: 27716480 Free PMC article. Review. - Essential tension and constructive destruction: the spindle checkpoint and its regulatory links with mitotic exit.
Tan AL, Rida PC, Surana U. Tan AL, et al. Biochem J. 2005 Feb 15;386(Pt 1):1-13. doi: 10.1042/BJ20041415. Biochem J. 2005. PMID: 15521820 Free PMC article. Review. - Meiosis-Specific Functions of Kinesin Motors in Cohesin Removal and Maintenance of Chromosome Integrity in Budding Yeast.
Mittal P, Ghule K, Trakroo D, Prajapati HK, Ghosh SK. Mittal P, et al. Mol Cell Biol. 2020 Mar 30;40(8):e00386-19. doi: 10.1128/MCB.00386-19. Print 2020 Mar 30. Mol Cell Biol. 2020. PMID: 31964755 Free PMC article. - Spindle assembly requires complete disassembly of spindle remnants from the previous cell cycle.
Woodruff JB, Drubin DG, Barnes G. Woodruff JB, et al. Mol Biol Cell. 2012 Jan;23(2):258-67. doi: 10.1091/mbc.E11-08-0701. Epub 2011 Nov 16. Mol Biol Cell. 2012. PMID: 22090343 Free PMC article. - Degradation of Ndd1 by APC/C(Cdh1) generates a feed forward loop that times mitotic protein accumulation.
Sajman J, Zenvirth D, Nitzan M, Margalit H, Simpson-Lavy KJ, Reiss Y, Cohen I, Ravid T, Brandeis M. Sajman J, et al. Nat Commun. 2015 May 11;6:7075. doi: 10.1038/ncomms8075. Nat Commun. 2015. PMID: 25959309
References
- Amon A, Irniger S, Nasmyth K. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell. 1994;77:1037–1050. - PubMed
- Antonio C, Ferby I, Wilhelm H, Jones M, Karsenti E, Nebreda AR, Vernos I. Xkid, a chromokinesin required for chromosome alignment on the metaphase plate. Cell. 2000;102:425–435. - PubMed
- Baumer M, Braus GH, Irniger S. Two different modes of cyclin clb2 proteolysis during mitosis in Saccharomyces cerevisiae. FEBS Lett. 2000a;468:142–148. - PubMed
- Baumer M, Kunzler M, Steigemann P, Braus GH, Irniger S. Yeast Ran-binding protein Yrb1p is required for efficient proteolysis of cell cycle regulatory proteins Pds1p, and Sic1p. J Biol Chem. 2000b;275:38929–38937. - PubMed
- Blangy A, Lane HA, d'Hérin P, Harper M, Kress M, Nigg EA. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 1995;83:1159–1169. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous