Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses - PubMed (original) (raw)
. 2001 Nov 20;98(24):13790-5.
doi: 10.1073/pnas.191502998. Epub 2001 Nov 13.
W G Richards, J Staunton, C Li, S Monti, P Vasa, C Ladd, J Beheshti, R Bueno, M Gillette, M Loda, G Weber, E J Mark, E S Lander, W Wong, B E Johnson, T R Golub, D J Sugarbaker, M Meyerson
Affiliations
- PMID: 11707567
- PMCID: PMC61120
- DOI: 10.1073/pnas.191502998
Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses
A Bhattacharjee et al. Proc Natl Acad Sci U S A. 2001.
Abstract
We have generated a molecular taxonomy of lung carcinoma, the leading cause of cancer death in the United States and worldwide. Using oligonucleotide microarrays, we analyzed mRNA expression levels corresponding to 12,600 transcript sequences in 186 lung tumor samples, including 139 adenocarcinomas resected from the lung. Hierarchical and probabilistic clustering of expression data defined distinct subclasses of lung adenocarcinoma. Among these were tumors with high relative expression of neuroendocrine genes and of type II pneumocyte genes, respectively. Retrospective analysis revealed a less favorable outcome for the adenocarcinomas with neuroendocrine gene expression. The diagnostic potential of expression profiling is emphasized by its ability to discriminate primary lung adenocarcinomas from metastases of extra-pulmonary origin. These results suggest that integration of expression profile data with clinical parameters could aid in diagnosis of lung cancer patients.
Figures
Figure 1
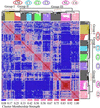
Hierarchical clustering defines subclasses of lung tumors. Two-dimensional hierarchical clustering of 203 lung tumors and normal lung samples was performed with 3,312 transcript sequences. The normalized expression index for each transcript sequence (rows) in each sample (columns) is indicated by a color code (see EXPRESSION INDEX bar at lower left of figure). (A) Clusters of genes with high relative expression in normal lung (NL, pink branch). (B) Neuroendocrine tumors: small-cell lung cancer (SCLC, gold branch) and pulmonary carcinoids (COID, light blue branch). (C) squamous cell lung carcinomas with keratin markers (SQ, light green branch). (D) Proliferation-related markers. Adenocarcinomas resected from the lung (black branches) and a subset of adenocarcinomas suspected as colon metastases (red branch) are indicated. Color bars on the right correspond to regions displayed in Fig. 10.
Figure 2
Clustering defines adenocarcinoma subclasses. Comparison of classifications derived by hierarchical clustering (dendrogram) and probabilistic clustering (colored matrix) algorithms. The two-dimensional colored matrix is a visual representation of a corresponding numerical matrix whose entries record a normalized measure of association strength between samples. Strong association approaches a value of 1 (red) and poor association is close to 0 (blue). CM, colon metastasis; NL, normal lung; C1 through C4 are adenocarcinoma clusters; I (gold), II, and III are additional groups with weaker association. See Figs. 12–15 for details and sample names.
Figure 3
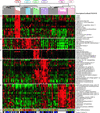
Gene expression clusters and histologic differentiation within lung adenocarcinoma subclasses. Genes expressed at high levels in specific subsets of adenocarcinomas. The normalized expression index is shown as in Fig. 1. (A) Colon metastases. (B) Proliferation-related gene expression (C1). (C) Neuroendocrine gene expression (C2). (D) Ornithine decarboxylase 1 and surfactant gene expression (C3 and C2). (E) Type II pneumocyte gene expression (C4, C3, and normal lung). (F) Histopathological degree of differentiation (red, poor; yellow, moderate; green, well; white, not available or irrelevant). (G) Estimated nucleated tumor content: white, not determined or irrelevant; gray, 30–40%; blue, 40–70%; black, greater than 70%).
Figure 4
Survival analysis of neuroendocrine C2 adenocarcinomas. Kaplan–Meier curves for C2 versus all other adenocarcinomas. (A) All patients: C2, n = 9; non-C2, n = 117. (B) Patients with stage I tumors only: C2, n = 4; non-C2, n = 72.
Similar articles
- Diversity of gene expression in adenocarcinoma of the lung.
Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen GD, Perou CM, Whyte RI, Altman RB, Brown PO, Botstein D, Petersen I. Garber ME, et al. Proc Natl Acad Sci U S A. 2001 Nov 20;98(24):13784-9. doi: 10.1073/pnas.241500798. Epub 2001 Nov 13. Proc Natl Acad Sci U S A. 2001. PMID: 11707590 Free PMC article. - Predicting survival within the lung cancer histopathological hierarchy using a multi-scale genomic model of development.
Liu H, Kho AT, Kohane IS, Sun Y. Liu H, et al. PLoS Med. 2006 Jul;3(7):e232. doi: 10.1371/journal.pmed.0030232. PLoS Med. 2006. PMID: 16800721 Free PMC article. - Smoking and cancer-related gene expression in bronchial epithelium and non-small-cell lung cancers.
Woenckhaus M, Klein-Hitpass L, Grepmeier U, Merk J, Pfeifer M, Wild P, Bettstetter M, Wuensch P, Blaszyk H, Hartmann A, Hofstaedter F, Dietmaier W. Woenckhaus M, et al. J Pathol. 2006 Oct;210(2):192-204. doi: 10.1002/path.2039. J Pathol. 2006. PMID: 16915569 - Gene expression profiling reveals reproducible human lung adenocarcinoma subtypes in multiple independent patient cohorts.
Hayes DN, Monti S, Parmigiani G, Gilks CB, Naoki K, Bhattacharjee A, Socinski MA, Perou C, Meyerson M. Hayes DN, et al. J Clin Oncol. 2006 Nov 1;24(31):5079-90. doi: 10.1200/JCO.2005.05.1748. J Clin Oncol. 2006. PMID: 17075127 Review. - [Clinicopathological study on primary lung cancer--immunohistochemical expression of p53 suppressor gene and bcl-2 oncogene in relation to prognosis].
Irie K, Ishida H, Furukawa T, Koyanagi K, Miyamoto Y. Irie K, et al. Rinsho Byori. 1996 Jan;44(1):32-41. Rinsho Byori. 1996. PMID: 8691638 Review. Japanese.
Cited by
- An Arf-Egr-C/EBPβ pathway linked to ras-induced senescence and cancer.
Salotti J, Sakchaisri K, Tourtellotte WG, Johnson PF. Salotti J, et al. Mol Cell Biol. 2015 Mar;35(5):866-83. doi: 10.1128/MCB.01489-14. Epub 2014 Dec 22. Mol Cell Biol. 2015. PMID: 25535333 Free PMC article. - Alterations in gene expression of proprotein convertases in human lung cancer have a limited number of scenarios.
Demidyuk IV, Shubin AV, Gasanov EV, Kurinov AM, Demkin VV, Vinogradova TV, Zinovyeva MV, Sass AV, Zborovskaya IB, Kostrov SV. Demidyuk IV, et al. PLoS One. 2013;8(2):e55752. doi: 10.1371/journal.pone.0055752. Epub 2013 Feb 7. PLoS One. 2013. PMID: 23409034 Free PMC article. - Non-negative matrix factorization by maximizing correntropy for cancer clustering.
Wang JJ, Wang X, Gao X. Wang JJ, et al. BMC Bioinformatics. 2013 Mar 24;14:107. doi: 10.1186/1471-2105-14-107. BMC Bioinformatics. 2013. PMID: 23522344 Free PMC article. - A miRNA-regulatory network explains how dysregulated miRNAs perturb oncogenic processes across diverse cancers.
Plaisier CL, Pan M, Baliga NS. Plaisier CL, et al. Genome Res. 2012 Nov;22(11):2302-14. doi: 10.1101/gr.133991.111. Epub 2012 Jun 28. Genome Res. 2012. PMID: 22745231 Free PMC article. - Selective requirement for Mediator MED23 in Ras-active lung cancer.
Yang X, Zhao M, Xia M, Liu Y, Yan J, Ji H, Wang G. Yang X, et al. Proc Natl Acad Sci U S A. 2012 Oct 9;109(41):E2813-22. doi: 10.1073/pnas.1204311109. Epub 2012 Sep 17. Proc Natl Acad Sci U S A. 2012. PMID: 22988093 Free PMC article.
References
- Greenlee R T, Hill-Harmon M B, Murray T, Thun M. CA Cancer J Clin. 2001;51:15–36. - PubMed
- Druker B J, Talpaz M, Resta D J, Peng B, Buchdunger E, Ford J M, Lydon N B, Kantarjian H, Capdeville R, Ohno-Jones S, et al. N Engl J Med. 2001;344:1031–1037. - PubMed
- Travis W D, Travis L B, Devesa S S. Cancer. 1995;75:191–202. - PubMed
- Sorensen J B, Hirsch F R, Gazdar A, Olsen J E. Cancer. 1993;71:2971–2976. - PubMed
- Breathnach O S, Kwiatkowski D J, Finkelstein D M, Godleski J, Sugarbaker D J, Johnson B E, Mentzer S. J Thorac Cardiovasc Surg. 2001;121:42–47. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases