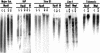Lsh, a member of the SNF2 family, is required for genome-wide methylation - PubMed (original) (raw)
Lsh, a member of the SNF2 family, is required for genome-wide methylation
K Dennis et al. Genes Dev. 2001.
Abstract
Methylation patterns of the mammalian genome are thought to be crucial for development. The precise mechanisms designating specific genomic loci for methylation are not known. Targeted deletion of Lsh results in perinatal lethality with a rather normal development. We report here, however, that Lsh(-/-) mice show substantial loss of methylation throughout the genome. The hypomethylated loci comprise repetitive elements and single copy genes. This suggests that global genomic methylation is not absolutely required for normal embryogenesis. Based on the similarity of Lsh to other SNF2 chromatin remodeling proteins, it suggests that alteration of chromatin affects global methylation patterns in mice.
Figures
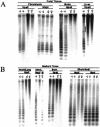
Figure 1
Hypomethylation of minor satellite sequences in _Lsh_−/− mice. (A) Southern analysis of genomic DNA derived at day 13.5 of gestation. DNA was digested with _Hpa_II or _Msp_I, blotted, and probed for minor satellite sequences using MR150. (B) Southern analysis of genomic DNA derived from newborn mice within 24 h after birth. Whole body comprises every tissue with the exception of the examined internal organs. DNA was digested with _Hpa_II or _Msp_I, blotted, and probed for minor satellite sequences using MR150.
Figure 2
Hypomethylation of repetitive sequences in _Lsh_−/− mice. Southern analysis for repetitive sequences. Genomic DNA derived from _Lsh_−/− or littermate controls was digested with _Mae_II and probed for major satellite sequences, or digested with _Hpa_II and _Msp_I (M) and probed for IAP, Sine B1, Line 1, or telomeric sequences.
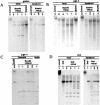
Figure 3
Hypomethylation of single copy sequences in _Lsh_−/− mice. (A) Southern analysis of the β-Globin gene. Genomic DNA was derived from newborn mice or embryos at day 13.5 gestation. DNA was digested with _Bam_HI with or without the methylation sensitive restriction enzyme _Hha_I, blotted, and probed for β-Globin. (B) Southern analysis of the Pgk-2 gene. (C) Southern analysis of the Pgk-1 gene. (D) Southern analysis of the H19 upstream imprinted region.
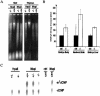
Figure 4
Global hypomethylation in _Lsh_−/− mice. (A) Genomic DNA from embryonal fibroblasts (MEF) or adult thymus from radiation chimeras (Geiman and Muegge 2000) was digested with the methylation-sensitive enzyme _Hpa_II and the nonsensitive enzyme _Msp_I, subjected to agarose gel electrophoresis, and visualized by ethidium bromide stain. (B) Methyl acceptance assay. Equal amounts of genomic DNA derived from day 13.5 embryonic body, embryonic liver, or from newborn brain were methylated in vitro by _Sss_I CG methylase using radiolabeled S-adenosyl-methionine as donor. This approach allows determination of the amount of unmethylated CG sites in the genome and serves as an indirect measurement of genomic methylation levels. The amount of incorporated radiolabeled methyl groups on cytosines per microgram of DNA was measured in Lsh deleted samples and control littermates as described previously (Antoun et al. 2000). (C) Direct measurement of methyl-cytosine in genomic DNA. Equal amounts of genomic DNA derived from brain samples of _Lsh_−/− mice and littermate controls were digested with _Msp_I, radiolabeled at the 5′-ends, and digested with nuclease P1 to generate 5′-deoxymononucleotides. Cytosine and methyl-cytosine were separated by thin layer chromatography (Cedar et al. 1979) and quantified using PhosphorImager analysis. The ratio of methyl-cytosine to total cytosine indicates the level of methylation at all CCGG sites. _Hpa_II digests should not generate methyl-cytosine spots and serve as controls, indicating the specificity of the assay.
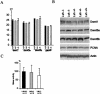
Figure 5
Expression of DNA methyltransferases and measurement of Mtase activity in _Lsh_−/− mice. (A) RT–PCR analysis. Total RNA of embryonic body (2 wild type, 1 heterozygote, and 3 knockout) derived from day 17.5 gestation was reverse transcribed and subjected to real-time PCR analysis for measurement of Dnmt1, Dnmt3a, or Dnmt3b or Gapdh transcripts as control. (CT) Cycle threshold, cycle number at which each PCR reaction reaches a predetermined fluorescence threshold, set within the linear range of all reactions. (B) Western analysis. Cellular extracts derived from fetal brain tissue were analyzed using specific antiserum against murine Dnmt1, Dnmt3a (Imgenex), Dnmt3b (Affinity Bioreagents), β-Actin (Sigma), or PCNA (Santa Cruz) as control. A similar result was obtained using lysates derived from embryonic body of day 17.5 gestation. (C) Mtase activity. Cellular extracts were prepared from indicated tissues and examined in vitro for their ability to transfer radio-labeled methyl-groups onto synthetic template poly[d(I–C)]·poly[d(I–C)] (Li et al. 1992). Embryonic bodies are from day 13.5 gestation.
Similar articles
- Lsh controls silencing of the imprinted Cdkn1c gene.
Fan T, Hagan JP, Kozlov SV, Stewart CL, Muegge K. Fan T, et al. Development. 2005 Feb;132(4):635-44. doi: 10.1242/dev.01612. Epub 2005 Jan 12. Development. 2005. PMID: 15647320 - Lsh, a SNF2 family member, is required for normal murine development.
Geiman TM, Tessarollo L, Anver MR, Kopp JB, Ward JM, Muegge K. Geiman TM, et al. Biochim Biophys Acta. 2001 May 3;1526(2):211-20. doi: 10.1016/s0304-4165(01)00129-5. Biochim Biophys Acta. 2001. PMID: 11325543 - Lsh, chromatin remodeling family member, modulates genome-wide cytosine methylation patterns at nonrepeat sequences.
Tao Y, Xi S, Shan J, Maunakea A, Che A, Briones V, Lee EY, Geiman T, Huang J, Stephens R, Leighty RM, Zhao K, Muegge K. Tao Y, et al. Proc Natl Acad Sci U S A. 2011 Apr 5;108(14):5626-31. doi: 10.1073/pnas.1017000108. Epub 2011 Mar 22. Proc Natl Acad Sci U S A. 2011. PMID: 21427231 Free PMC article. - Helicase homologues maintain cytosine methylation in plants and mammals.
Bourc'his D, Bestor TH. Bourc'his D, et al. Bioessays. 2002 Apr;24(4):297-9. doi: 10.1002/bies.10078. Bioessays. 2002. PMID: 11948614 Review. - Lsh, a guardian of heterochromatin at repeat elements.
Muegge K. Muegge K. Biochem Cell Biol. 2005 Aug;83(4):548-54. doi: 10.1139/o05-119. Biochem Cell Biol. 2005. PMID: 16094458 Review.
Cited by
- The ICF syndrome protein CDCA7 harbors a unique DNA binding domain that recognizes a CpG dyad in the context of a non-B DNA.
Hardikar S, Ren R, Ying Z, Zhou J, Horton JR, Bramble MD, Liu B, Lu Y, Liu B, Coletta LD, Shen J, Dan J, Zhang X, Cheng X, Chen T. Hardikar S, et al. Sci Adv. 2024 Aug 23;10(34):eadr0036. doi: 10.1126/sciadv.adr0036. Epub 2024 Aug 23. Sci Adv. 2024. PMID: 39178265 Free PMC article. - The SNF2 family ATPase LSH promotes cell-autonomous de novo DNA methylation in somatic cells.
Termanis A, Torrea N, Culley J, Kerr A, Ramsahoye B, Stancheva I. Termanis A, et al. Nucleic Acids Res. 2016 Sep 19;44(16):7592-604. doi: 10.1093/nar/gkw424. Epub 2016 May 13. Nucleic Acids Res. 2016. PMID: 27179028 Free PMC article. - Chromatin remodeling of histone H3 variants underlies epigenetic inheritance of DNA methylation.
Lee SC, Adams DW, Ipsaro JJ, Cahn J, Lynn J, Kim HS, Berube B, Major V, Calarco JP, LeBlanc C, Bhattacharjee S, Ramu U, Grimanelli D, Jacob Y, Voigt P, Joshua-Tor L, Martienssen RA. Lee SC, et al. bioRxiv [Preprint]. 2023 Aug 2:2023.07.11.548598. doi: 10.1101/2023.07.11.548598. bioRxiv. 2023. PMID: 37503143 Free PMC article. Updated. Preprint. - Epigenetic regulation of the stem cell mitogen Fgf-2 by Mbd1 in adult neural stem/progenitor cells.
Li X, Barkho BZ, Luo Y, Smrt RD, Santistevan NJ, Liu C, Kuwabara T, Gage FH, Zhao X. Li X, et al. J Biol Chem. 2008 Oct 10;283(41):27644-27652. doi: 10.1074/jbc.M804899200. Epub 2008 Aug 8. J Biol Chem. 2008. PMID: 18689796 Free PMC article. - Dnmt3b Prefers Germ Line Genes and Centromeric Regions: Lessons from the ICF Syndrome and Cancer and Implications for Diseases.
Walton EL, Francastel C, Velasco G. Walton EL, et al. Biology (Basel). 2014 Sep 5;3(3):578-605. doi: 10.3390/biology3030578. Biology (Basel). 2014. PMID: 25198254 Free PMC article. Review.
References
- Antoun G, Baylin SB, Ali-Osman F. DNA methyltransferase levels and altered CpG methylation in the total genome and in the GSTP1 gene in human glioma cells transfected with sense and antisense DNA methyltransferase cDNA. J Cell Biochem. 2000;77:372–381. - PubMed
- Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: Epigenetics joins genetics. Trends Genet. 2000;16:168–174. - PubMed
- Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases