Non-coding RNAs: the architects of eukaryotic complexity - PubMed (original) (raw)
Non-coding RNAs: the architects of eukaryotic complexity
J S Mattick. EMBO Rep. 2001 Nov.
Abstract
Around 98% of all transcriptional output in humans is non-coding RNA. RNA-mediated gene regulation is widespread in higher eukaryotes and complex genetic phenomena like RNA interference, co-suppression, transgene silencing, imprinting, methylation, and possibly position-effect variegation and transvection, all involve intersecting pathways based on or connected to RNA signaling. I suggest that the central dogma is incomplete, and that intronic and other non-coding RNAs have evolved to comprise a second tier of gene expression in eukaryotes, which enables the integration and networking of complex suites of gene activity. Although proteins are the fundamental effectors of cellular function, the basis of eukaryotic complexity and phenotypic variation may lie primarily in a control architecture composed of a highly parallel system of trans-acting RNAs that relay state information required for the coordination and modulation of gene expression, via chromatin remodeling, RNA-DNA, RNA-RNA and RNA-protein interactions. This system has interesting and perhaps informative analogies with small world networks and dataflow computing.
Figures
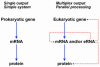
Fig. 1. Comparison of the prokaryotic and proposed eukaryotic genetic operating systems. The left panel shows the central dogma in which genes code, via mRNA, for proteins, which carry out the catalytic, structural, signal transduction and regulatory functions of the cell. The right panel shows the proposed operating system in eukaryotes wherein genes may express two levels of information: mRNA for proteins, and eRNAs that carry out concomitant networking and other functions within the organism. Thus there are three types of genes in eukaryotes: those that encode only protein (which are rare), those that encode only eRNA, and those that encode both.
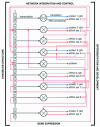
Fig. 2. A more detailed schematic of the proposed role of eRNAs in eukaryotic system networking and control. Genes, packaged in chromatin, express primary transcripts which are then (alternatively) spliced to yield an mRNA and/or n introns, which may be further processed to form multiple smaller species, such as let-7. Some noncoding RNA genes may yield functional RNAs from both introns and exons (nRNA). These RNAs may then act as signaling or guide molecules to integrate activity at this locus with that of related parts of the network, via effects on chromatin structure, transcription, splicing, other levels of RNA processing, mRNA translation, mRNA stability and other levels of RNA-mediated signal transduction within the cell. The evidence indicates that many if not most of these interactions will be homology (primary sequence) dependent, and involve RNA–DNA, RNA–RNA and RNA–protein interactions, but others may involve secondary or tertiary RNA structures and RNA-mediated catalysis. This scheme is not comprehensive, but is intended to give a sense of the complexity and potential of such networks for programmed control and system integration of complex suites of gene activity in differentiation and development.
John S. Mattick
Similar articles
- The evolution of controlled multitasked gene networks: the role of introns and other noncoding RNAs in the development of complex organisms.
Mattick JS, Gagen MJ. Mattick JS, et al. Mol Biol Evol. 2001 Sep;18(9):1611-30. doi: 10.1093/oxfordjournals.molbev.a003951. Mol Biol Evol. 2001. PMID: 11504843 Review. - Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms.
Mattick JS. Mattick JS. Bioessays. 2003 Oct;25(10):930-9. doi: 10.1002/bies.10332. Bioessays. 2003. PMID: 14505360 Review. - The relationship between non-protein-coding DNA and eukaryotic complexity.
Taft RJ, Pheasant M, Mattick JS. Taft RJ, et al. Bioessays. 2007 Mar;29(3):288-99. doi: 10.1002/bies.20544. Bioessays. 2007. PMID: 17295292 - As antisense RNA gets intronic.
Reis EM, Louro R, Nakaya HI, Verjovski-Almeida S. Reis EM, et al. OMICS. 2005 Spring;9(1):2-12. doi: 10.1089/omi.2005.9.2. OMICS. 2005. PMID: 15805775 Review. - The RNA infrastructure: an introduction to ncRNA networks.
Collins LJ. Collins LJ. Adv Exp Med Biol. 2011;722:1-19. doi: 10.1007/978-1-4614-0332-6_1. Adv Exp Med Biol. 2011. PMID: 21915779 Review.
Cited by
- Profiling of mRNA and long non-coding RNA of urothelial cancer in recipients after renal transplantation.
Shang D, Zheng T, Zhang J, Tian Y, Liu Y. Shang D, et al. Tumour Biol. 2016 Sep;37(9):12673-12684. doi: 10.1007/s13277-016-5148-1. Epub 2016 Jul 22. Tumour Biol. 2016. PMID: 27448299 - Unveiling the Network regulatory mechanism of ncRNAs on the Ferroptosis Pathway: Implications for Preeclampsia.
Zhang Y, Zhang J, Chen S, Li M, Yang J, Tan J, He B, Zhu L. Zhang Y, et al. Int J Womens Health. 2024 Sep 30;16:1633-1651. doi: 10.2147/IJWH.S485653. eCollection 2024. Int J Womens Health. 2024. PMID: 39372667 Free PMC article. Review. - Role of microRNAs in type 2 diseases and allergen-specific immunotherapy.
Jakwerth CA, Kitzberger H, Pogorelov D, Müller A, Blank S, Schmidt-Weber CB, Zissler UM. Jakwerth CA, et al. Front Allergy. 2022 Sep 12;3:993937. doi: 10.3389/falgy.2022.993937. eCollection 2022. Front Allergy. 2022. PMID: 36172292 Free PMC article. Review. - Searching for convergent pathways in autism spectrum disorders: insights from human brain transcriptome studies.
Gokoolparsadh A, Sutton GJ, Charamko A, Green NF, Pardy CJ, Voineagu I. Gokoolparsadh A, et al. Cell Mol Life Sci. 2016 Dec;73(23):4517-4530. doi: 10.1007/s00018-016-2304-0. Epub 2016 Jul 12. Cell Mol Life Sci. 2016. PMID: 27405608 Free PMC article. Review. - Intron size and exon evolution in Drosophila.
Marais G, Nouvellet P, Keightley PD, Charlesworth B. Marais G, et al. Genetics. 2005 May;170(1):481-5. doi: 10.1534/genetics.104.037333. Epub 2005 Mar 21. Genetics. 2005. PMID: 15781704 Free PMC article.
References
- Akhtar A., Zink, D. and Becker, P.B. (2000) Chromodomains are protein–RNA interaction modules. Nature, 407, 405–409. - PubMed
- Albert R., Jeong, H. and Barabasi, A.L. (2000) Error and attack tolerance of complex networks. Nature, 406, 378–382. - PubMed
- Askew D.S. and Xu, F. (1999) New insights into the function of noncoding RNA and its potential role in disease pathogenesis. Histol. Histopathol., 14, 235–241. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources