RNAMotif, an RNA secondary structure definition and search algorithm - PubMed (original) (raw)
RNAMotif, an RNA secondary structure definition and search algorithm
T J Macke et al. Nucleic Acids Res. 2001.
Abstract
RNA molecules fold into characteristic secondary and tertiary structures that account for their diverse functional activities. Many of these RNA structures are assembled from a collection of RNA structural motifs. These basic building blocks are used repeatedly, and in various combinations, to form different RNA types and define their unique structural and functional properties. Identification of recurring RNA structural motifs will therefore enhance our understanding of RNA structure and help associate elements of RNA structure with functional and regulatory elements. Our goal was to develop a computer program that can describe an RNA structural element of any complexity and then search any nucleotide sequence database, including the complete prokaryotic and eukaryotic genomes, for these structural elements. Here we describe in detail a new computational motif search algorithm, RNAMotif, and demonstrate its utility with some motif search examples. RNAMotif differs from other motif search tools in two important aspects: first, the structure definition language is more flexible and can specify any type of base-base interaction; second, RNAMotif provides a user controlled scoring section that can be used to add capabilities that patterns alone cannot provide.
Figures
Figure 1
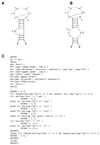
Examples of RNA structure motifs and descriptor constraints with important conserved nucleotides and scoring values. (A) A helical stem closed by a tetraloop. (B) An E-loop motif. (C) The core of the E-loop depicted with the observed non-canonical base pairing interactions. The most significant structural elements within the motif are shown within the dotted box. The structural components (stems or single-stranded regions) that flank this internal loop can vary in length and composition. (D) E-loop descriptor derived from the constraints shown in (B). Additional constraints for the stems flanking the core motif not shown were used in the overall score calculations reported in Table 2.
Figure 1
Examples of RNA structure motifs and descriptor constraints with important conserved nucleotides and scoring values. (A) A helical stem closed by a tetraloop. (B) An E-loop motif. (C) The core of the E-loop depicted with the observed non-canonical base pairing interactions. The most significant structural elements within the motif are shown within the dotted box. The structural components (stems or single-stranded regions) that flank this internal loop can vary in length and composition. (D) E-loop descriptor derived from the constraints shown in (B). Additional constraints for the stems flanking the core motif not shown were used in the overall score calculations reported in Table 2.
Figure 1
Examples of RNA structure motifs and descriptor constraints with important conserved nucleotides and scoring values. (A) A helical stem closed by a tetraloop. (B) An E-loop motif. (C) The core of the E-loop depicted with the observed non-canonical base pairing interactions. The most significant structural elements within the motif are shown within the dotted box. The structural components (stems or single-stranded regions) that flank this internal loop can vary in length and composition. (D) E-loop descriptor derived from the constraints shown in (B). Additional constraints for the stems flanking the core motif not shown were used in the overall score calculations reported in Table 2.
Figure 2
Models for the IRE structure and generic descriptor. (A) IRE bulge model. (B) IRE internal loop model. (C) Descriptor that captures both types of IRE.
Figure 3
SRP-RNA alignment and structure. (A) Structure-based alignment of the domain IV stem–loop region of SRP-RNA based on the full-length alignment from the SRP-RNA website (http://psyche.uthct.edu/dbs/SRPDB/SRPDB.html). Sequences of a few representative organisms from the three major phylogenetic kingdoms (Bacteria, Archaea and Eukarya) are shown here. The top two lines of the alignment show the base pairing schema. ( and ) are used to denote the 5′ and 3′ sides of a helix, respectively, and the numbers indicate the paired segments. (B) A consensus diagram of the SRP-RNA structure derived from analyzing the alignment of over 100 organisms. The biases in nucleotides and range of size variations in loops and helices are shown. An RNAMotif descriptor (not shown) was derived based on the constraints shown here.
Figure 4
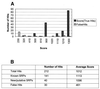
Summary of results of a RNAMotif search for SRP-RNA. (A) Score distribution of true and false hits based on RNAMotif scores in a search of the GenBank nucleotide database with an SRP-RNA descriptor. (B) Summary of scores for true and false hits.
Similar articles
- Escherichia coli DbpA is a 3' --> 5' RNA helicase.
Diges CM, Uhlenbeck OC. Diges CM, et al. Biochemistry. 2005 May 31;44(21):7903-11. doi: 10.1021/bi050033x. Biochemistry. 2005. PMID: 15910005 - Secondary structure prediction for aligned RNA sequences.
Hofacker IL, Fekete M, Stadler PF. Hofacker IL, et al. J Mol Biol. 2002 Jun 21;319(5):1059-66. doi: 10.1016/S0022-2836(02)00308-X. J Mol Biol. 2002. PMID: 12079347 - Characterization of transient RNA-RNA interactions important for the facilitated structure formation of bacterial ribosomal 16S RNA.
Besançon W, Wagner R. Besançon W, et al. Nucleic Acids Res. 1999 Nov 15;27(22):4353-62. doi: 10.1093/nar/27.22.4353. Nucleic Acids Res. 1999. PMID: 10536142 Free PMC article. - 5'- and 3'-noncoding regions in flavivirus RNA.
Markoff L. Markoff L. Adv Virus Res. 2003;59:177-228. doi: 10.1016/s0065-3527(03)59006-6. Adv Virus Res. 2003. PMID: 14696330 Free PMC article. Review. - [Structure and function of the non-coding regions of hepatitis C viral RNA].
Dutkiewicz M, Swiqtkowska A, Ciesiołka J. Dutkiewicz M, et al. Postepy Biochem. 2006;52(1):62-71. Postepy Biochem. 2006. PMID: 16869303 Review. Polish.
Cited by
- A universal RNA structural motif docking the elbow of tRNA in the ribosome, RNAse P and T-box leaders.
Lehmann J, Jossinet F, Gautheret D. Lehmann J, et al. Nucleic Acids Res. 2013 May 1;41(10):5494-502. doi: 10.1093/nar/gkt219. Epub 2013 Apr 10. Nucleic Acids Res. 2013. PMID: 23580544 Free PMC article. - Genome-wide screen and functional analysis in Xanthomonas reveal a large number of mRNA-derived sRNAs, including the novel RsmA-sequester RsmU.
Tang DJ, Chen XL, Jia Y, Liang YW, He YP, Lu TT, Zhu CR, Han B, An SQ, Tang JL. Tang DJ, et al. Mol Plant Pathol. 2020 Dec;21(12):1573-1590. doi: 10.1111/mpp.12997. Epub 2020 Sep 23. Mol Plant Pathol. 2020. PMID: 32969159 Free PMC article. - Query-dependent banding (QDB) for faster RNA similarity searches.
Nawrocki EP, Eddy SR. Nawrocki EP, et al. PLoS Comput Biol. 2007 Mar 30;3(3):e56. doi: 10.1371/journal.pcbi.0030056. Epub 2007 Feb 7. PLoS Comput Biol. 2007. PMID: 17397253 Free PMC article. - RNA localization signals: deciphering the message with bioinformatics.
Hamilton RS, Davis I. Hamilton RS, et al. Semin Cell Dev Biol. 2007 Apr;18(2):178-85. doi: 10.1016/j.semcdb.2007.02.001. Epub 2007 Feb 12. Semin Cell Dev Biol. 2007. PMID: 17452113 Free PMC article. Review. - Inverse folding based pre-training for the reliable identification of intrinsic transcription terminators.
Brandenburg VB, Narberhaus F, Mosig A. Brandenburg VB, et al. PLoS Comput Biol. 2022 Jul 7;18(7):e1010240. doi: 10.1371/journal.pcbi.1010240. eCollection 2022 Jul. PLoS Comput Biol. 2022. PMID: 35797361 Free PMC article.
References
- Theil E.C. (1998) The iron responsive element (IRE) family of mRNA regulators. Regulation of iron transport and uptake compared in animals, plants and microorganisms. Met. Ions Biol. Syst., 35, 403–434. - PubMed
- Kim H.Y., LaVaute,T., Iwai,K., Klausner,R.D. and Rouault,T.A. (1996) Identification of a conserved and functional iron-responsive element in the 5′-untranslated region of mammalian mitochondrial aconitase. J. Biol. Chem., 271, 24226–24230. - PubMed
- Son S.Y. (1993) The structure and regulation of histone genes. Saenghwahak Nyusu, 13, 64–70.
- Shepherd R.K., Gabryszuk,J., Al-Ali,M., Allen,C.A., Joyce,I., Holmes,W.M. and Zehner,Z.E. (1997) A dual stem–loop structure in the 3′ untranslated region of vimentin mRNA binds specific protein(s). Nucleic Acids Symp. Ser., 36, 142–145.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources