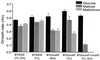New regulatory gene that contributes to control of Bacteroides thetaiotaomicron starch utilization genes - PubMed (original) (raw)
New regulatory gene that contributes to control of Bacteroides thetaiotaomicron starch utilization genes
K H Cho et al. J Bacteriol. 2001 Dec.
Abstract
Bacteroides thetaiotaomicron uses starch as a source of carbon and energy. Early steps in the pathway of starch utilization, such as starch binding and starch hydrolysis, are encoded by sus genes, which have been characterized previously. The sus structural genes are expressed only if cells are grown in medium containing maltose or higher oligomers of glucose. Regulation of the sus structural genes is mediated by SusR, an activator that is encoded by a gene located next to the sus structural genes. A strain with a disruption in susR cannot grow on starch but can still grow on maltose and maltotriose. A search for transposon-generated mutants that could not grow on maltose and maltotriose unexpectedly located a gene, designated malR, which regulates expression of an alpha-glucosidase not controlled by SusR. Although a disruption in susR did not affect expression of the malR controlled gene, a disruption in malR reduced expression of the sus structural genes. Thus, MalR appears to participate with SusR in regulation of the sus genes. Results of transcriptional fusion assays and reverse transcription-PCR experiments showed that malR is expressed constitutively. Moreover, multiple copies of malR provided on a plasmid (5 to 10 copies per cell) more than doubled the amount of alpha-glucosidase activity in cell extracts. Our results demonstrate that the starch utilization system of B. thetaiotaomicron is controlled on at least two levels by the regulatory proteins SusR and MalR.
Figures
FIG. 1
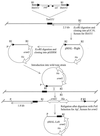
Cloning of pMAL-Right and pMAL-Left to sequence the region where Tn_4351_ is inserted in BTMAL. The position of the transposon insertion in BTMAL is indicated by the black rectangle. The heavy horizontal arrows in Tn_4351_ indicate the direct repeat insertion sequence (IS_4351_). Tn_4351_ carries an erythromycin resistance gene (ermF), which is expressed only in Bacteroides strains, and a tetracycline resistance gene (tetX), which is expressed only in aerobically grown E. coli strains. pGERM, a suicide vector in Bacteroides, has a different erythromycin resistance gene (ermG). The double lines (=) indicate Bacteroides chromosomal DNA. Abbreviations: RI, _Eco_RI; P, _Pst_I
FIG. 2
(A) Map showing the relative locations of ORFs in the malR gene area. The position of the transposon insertion in mutant BTMAL is indicated by the vertical arrow above the map. DNA segments used to make insertional disruptions are shown as horizontal lines under the map and marked with an “Ω.” The sizes of the DNA fragments used to make the disruptions are also indicated under the lines. gs, putative glutamine synthetase gene; malR, putative regulatory protein gene; β-nagA, putative β-_N_-acetylglucosaminidase gene. (B) Deduced amino acid sequence of malR. A possible carboxy-terminal helix-turn-helix motif is underlined. The vertical arrow indicates the transposon insertion site in the transposon-generated mutant, BTMAL.
FIG. 3
Growth rates of various mutants on glucose, maltose, and maltotriose. The medium used was defined medium that contained glucose, maltose, and maltotriose, and the results are indicated by the three shaded bars in each set from left to right, respectively. The concentrations of antibiotics used in this experiment were 3 μg/ml for tetracycline (Tc) and 10 μg/ml for erythromycin (Em). These measurements were done in triplicate; the range of values is indicated by the error bars. B. thetaiotaomicron BT4009 was used as the wild-type control because it contains a single copy of tetQ and ermF. Thus, either tetracycline or erythromycin or both (Tc, Em) can be added to the medium used to grow both the control and the mutant strains. This eliminates the slight differences in growth rate that can sometimes occur due to the presence of antibiotics in the medium (note the difference between “Tc” columns versus the “Tc, Em” columns). The selectable marker used to create the BTΩ_malR_ strain was ermF, and the marker used to create the BTΩ_susR_ strain was tetQ. The double mutant BTΩ_susR_Ω_malR_ contained both resistance genes.
FIG. 4
Detection of malR expression on glucose and maltose by using RT-PCR. Odd-numbered lanes contain negative controls in which the reaction was done without the RT step. The expression of the maltose-regulated susD gene was used as a control to assess whether we could detect regulated gene expression (lane 2, glucose [G]-grown cells; lane 4, maltose [M]-grown cells). The region amplified from susD was 0.5 kb in size. To check malR expression (lanes 6 and 8), a 0.4-kbp region of the mRNA was amplified. The 1-kb ladder (Gibco-BRL) is seen in the two outside lanes.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources