Underediting of glutamate receptor GluR-B mRNA in malignant gliomas - PubMed (original) (raw)
Underediting of glutamate receptor GluR-B mRNA in malignant gliomas
S Maas et al. Proc Natl Acad Sci U S A. 2001.
Abstract
In mammals, RNA editing by site-selective adenosine deamination regulates key functional properties of neurotransmitter receptors in the central nervous system. Glutamate receptor subunit B is nearly 100% edited at one position (the Q/R-site), which is essential for normal receptor function. Its significance is apparent from mouse models in which a slightly reduced rate of Q/R-site editing is associated with early onset epilepsy and premature death. Here we report that in tissues from malignant human brain tumors, this editing position of glutamate receptor subunit B is substantially underedited compared with control tissues. We also observe alterations in editing and alternative splicing of serotonin receptor 5-HT(2C) transcripts. These changes correlate with a decrease in enzymatic activity of the editing enzyme adenosine deaminase acting on RNA (ADAR) 2, as deduced from analysis of ADAR2 self-editing. Our results suggest a role for RNA editing in tumor progression and may provide a molecular model explaining the occurrence of epileptic seizures in association with malignant gliomas.
Figures
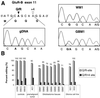
Figure 1
GluR-B Q/R site editing in human brain and gliomas. (A) Direct sequencing of GluR-B specific PCR amplicons from human genomic DNA (gDNA), reverse-transcribed RNA from white matter of human brain (WM1), and glioblastoma tissue (GBM1). Electropherograms covering the Q/R editing position within exon 11 are shown. Mixed sequence signals (A and G) indicate RNA editing, and the ratios of the peak heights reflect the extent of Q/R site editing. (B) Quantitation of editing of the Q/R site and Q/R +4 position in human control brain from gray matter (cortex) and white matter (WM A, WM B) in comparison to values from one low-grade human astrocytoma (AC1), various high-grade glioblastomas (GBM1–7), and glioma-derived cell lines GC1 and GC2. Percent values for examined samples, each derived from direct sequencing of at least three independent amplifications, poisoned primer extension analysis as well as subsequent sequence analysis of at least 50 individually subcloned cDNAs.
Figure 2
(A) Editing and alternative splicing of 5HT2C receptor pre-mRNA. Schematic representation of the serotonin receptor 5HT2C pre-mRNA, with the positions of the major sites for RNA editing (A–E) depicted. The positions of alternative splicing affecting the same region are indicated. Splicing of intron 3 leads to the long splice variant (LF), resulting in a functional receptor including the edited residues. Alternative splicing with partial deletion of exon 3 gives rise to the short splice form (SF), which is translated to yield an inactive receptor molecule and also lacks the edited sites. (B) Ratios of the 5HT2C splice forms in control human brain (cortex, WM A) versus glioma tissues (GBM1, 2, 6) and glioma-derived cell lines (GC1, 2). Percent values for the truncated variant (SF) and functional variant (LF) are determined by semiquantitative RT-PCR.
Figure 3
ADAR2 self-editing as an ADAR2 activity marker. (A) Intronic editing of ADAR2 pre-mRNA and resulting splice products. A-to-I editing at the −1 position by ADAR2 leads to partial inclusion of intron1. Only the regular splice form (Reg) results in a functional enzyme. (B) Characterization of stably transformed human embryonic kidney (HEK)293 cell lines expressing increasing amounts of the editing enzyme ADAR1 (N5, N8, N6) or ADAR2 (R10, R14, R13). Immunoblot analysis of cellular extracts from stable cell lines documents different ADAR expression levels. The relative adenosine deaminase activities measured in vitro on extended double-stranded RNA of the extracts (normalized to the activity of extract from native HEK 293 cells) are indicated. (C) Analysis of ADAR2 editing in ADAR-expressing cell lines. The percent values for the alternative splice form of ADAR2 resulting from ADAR2 pre-mRNA editing are given as measured by semiquantitative RT-PCR.
Similar articles
- Adenosine Deaminase That Acts on RNA 3 (ADAR3) Binding to Glutamate Receptor Subunit B Pre-mRNA Inhibits RNA Editing in Glioblastoma.
Oakes E, Anderson A, Cohen-Gadol A, Hundley HA. Oakes E, et al. J Biol Chem. 2017 Mar 10;292(10):4326-4335. doi: 10.1074/jbc.M117.779868. Epub 2017 Feb 6. J Biol Chem. 2017. PMID: 28167531 Free PMC article. - Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2.
Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH. Higuchi M, et al. Nature. 2000 Jul 6;406(6791):78-81. doi: 10.1038/35017558. Nature. 2000. PMID: 10894545 - Aberrant alternative splicing pattern of ADAR2 downregulates adenosine-to-inosine editing in glioma.
Li Z, Tian Y, Tian N, Zhao X, Du C, Han L, Zhang H. Li Z, et al. Oncol Rep. 2015 Jun;33(6):2845-52. doi: 10.3892/or.2015.3907. Epub 2015 Apr 8. Oncol Rep. 2015. PMID: 25873329 - The molecular link between inefficient GluA2 Q/R site-RNA editing and TDP-43 pathology in motor neurons of sporadic amyotrophic lateral sclerosis patients.
Yamashita T, Kwak S. Yamashita T, et al. Brain Res. 2014 Oct 10;1584:28-38. doi: 10.1016/j.brainres.2013.12.011. Epub 2013 Dec 16. Brain Res. 2014. PMID: 24355598 Review. - Novel etiological and therapeutic strategies for neurodiseases: RNA editing enzyme abnormality in sporadic amyotrophic lateral sclerosis.
Hideyama T, Yamashita T, Nishimoto Y, Suzuki T, Kwak S. Hideyama T, et al. J Pharmacol Sci. 2010;113(1):9-13. doi: 10.1254/jphs.09r21fm. Epub 2010 Apr 16. J Pharmacol Sci. 2010. PMID: 20424386 Review.
Cited by
- Modulation of microRNA editing, expression and processing by ADAR2 deaminase in glioblastoma.
Tomaselli S, Galeano F, Alon S, Raho S, Galardi S, Polito VA, Presutti C, Vincenti S, Eisenberg E, Locatelli F, Gallo A. Tomaselli S, et al. Genome Biol. 2015 Jan 13;16(1):5. doi: 10.1186/s13059-014-0575-z. Genome Biol. 2015. PMID: 25582055 Free PMC article. - Letter from the editor: Adenosine-to-inosine RNA editing in Alu repeats in the human genome.
Levanon K, Eisenberg E, Rechavi G, Levanon EY. Levanon K, et al. EMBO Rep. 2005 Sep;6(9):831-5. doi: 10.1038/sj.embor.7400507. EMBO Rep. 2005. PMID: 16138094 Free PMC article. Review. - Abnormal expression of an ADAR2 alternative splicing variant in gliomas downregulates adenosine-to-inosine RNA editing.
Wei J, Li Z, Du C, Qi B, Zhao X, Wang L, Bi L, Wang G, Zhang X, Su X, Pan Y, Tian Y. Wei J, et al. Acta Neurochir (Wien). 2014 Jun;156(6):1135-42. doi: 10.1007/s00701-014-2004-1. Epub 2014 Feb 11. Acta Neurochir (Wien). 2014. PMID: 24509948 Free PMC article. - Principles Governing A-to-I RNA Editing in the Breast Cancer Transcriptome.
Fumagalli D, Gacquer D, Rothé F, Lefort A, Libert F, Brown D, Kheddoumi N, Shlien A, Konopka T, Salgado R, Larsimont D, Polyak K, Willard-Gallo K, Desmedt C, Piccart M, Abramowicz M, Campbell PJ, Sotiriou C, Detours V. Fumagalli D, et al. Cell Rep. 2015 Oct 13;13(2):277-89. doi: 10.1016/j.celrep.2015.09.032. Epub 2015 Oct 1. Cell Rep. 2015. PMID: 26440892 Free PMC article. - ADAR2 functions as a tumor suppressor via editing IGFBP7 in esophageal squamous cell carcinoma.
Chen YB, Liao XY, Zhang JB, Wang F, Qin HD, Zhang L, Shugart YY, Zeng YX, Jia WH. Chen YB, et al. Int J Oncol. 2017 Feb;50(2):622-630. doi: 10.3892/ijo.2016.3823. Epub 2016 Dec 29. Int J Oncol. 2017. PMID: 28035363 Free PMC article.
References
- Maas S, Rich A. BioEssays. 2000;22:790–802. - PubMed
- Gerber A P, Keller W. Trends Biochem Sci. 2001;26:376–384. - PubMed
- Bass B L. Trends Biochem Sci. 1997;22:157–162. - PubMed
- Rueter S M, Emeson R B. In: Modification and Editing of RNA. Grosjean H, Benne R, editors. Washington, DC: Am. Soc. Microbiol.; 1998. pp. 343–361.
- Seeburg P H, Higuchi M, Sprengel R. Brain Res Brain Res Rev. 1998;26:217–229. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials