The intein of the Thermoplasma A-ATPase A subunit: structure, evolution and expression in E. coli - PubMed (original) (raw)
The intein of the Thermoplasma A-ATPase A subunit: structure, evolution and expression in E. coli
A G Senejani et al. BMC Biochem. 2001.
Abstract
Background: Inteins are selfish genetic elements that excise themselves from the host protein during post translational processing, and religate the host protein with a peptide bond. In addition to this splicing activity, most reported inteins also contain an endonuclease domain that is important in intein propagation.
Results: The gene encoding the Thermoplasma acidophilum A-ATPase catalytic subunit A is the only one in the entire T. acidophilum genome that has been identified to contain an intein. This intein is inserted in the same position as the inteins found in the ATPase A-subunits encoding gene in Pyrococcus abyssi, P. furiosus and P. horikoshii and is found 20 amino acids upstream of the intein in the homologous vma-1 gene in Saccharomyces cerevisiae. In contrast to the other inteins in catalytic ATPase subunits, the T. acidophilum intein does not contain an endonuclease domain.T. acidophilum has different codon usage frequencies as compared to Escherichia coli. Initially, the low abundance of rare tRNAs prevented expression of the T. acidophilum A-ATPase A subunit in E. coli. Using a strain of E. coli that expresses additional tRNAs for rare codons, the T. acidophilum A-ATPase A subunit was successfully expressed in E. coli.
Conclusions: Despite differences in pH and temperature between the E. coli and the T. acidophilum cytoplasms, the T. acidophilum intein retains efficient self-splicing activity when expressed in E. coli. The small intein in the Thermoplasma A-ATPase is closely related to the endonuclease containing intein in the Pyrococcus A-ATPase. Phylogenetic analyses suggest that this intein was horizontally transferred between Pyrococcus and Thermoplasma, and that the small intein has persisted in Thermoplasma apparently without homing.
Figures
Figure 1
Alignment of archaeal ATPase A-subunit intein sequences. The large gap in the Thermoplasma sequences indicates that the Thermoplasma acidophilum (Tac) and Thermoplasma volcanium (Tvo) inteins do not contain an endonuclease domain and only consist only of the self splicing domain, while the Pyrococcus abyssi (Pab), P. furiosus (Pfu) and P. horikoshii (Pho) A-ATPase A-subunits inteins are bifunctional. Regions corresponding to conserved blocks [18] are indicated in bold and labeled A-H. Blocks A, B, F and G are part of the splicing domain, whereas blocks C, D, E, H are typical for endonucleases of the LAGDIDAG type [18].
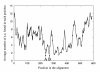
Figure 2
Divergence along the archaeal and vacuolar ATPase A subunits. The diagram depicts the number of substitutions per site calculated for a sliding window of 10 amino acid residues along an alignment of 62 vacuolar and archaeal ATPase catalytic subunits. The ordinate gives the number of substitutions observed in each position in the alignment. Arrows indicate the locations of the inteins. The first (A) gives the location of the inteins in Thermoplasma and Pyrococcus, the second (B) gives the location in the yeasts' vacuolar ATPases.
Figure 3
Comparison of vacuolar/archaeal ATPase A subunit and small subunit ribosomal RNA phylogenies. The phylogeny depicted in (A) was calculated from the extein portions of the genes only; (B) was calculated from small subunit ribosomal RNAs. The trees were calculated as unrooted, but are depicted as rooted using the eukaryotes as an outgroup to the Archaea (A), or the Crenarchaeota as an outgroup to the Euryarchaeota (B). Numbers give bootstrap values calculated from 100 bootstrapped samples analyzed using parsimony analysis. Values are only given for groups with more than 50% support. Asterisks denote branches that in the maximum likelihood evaluation were not at least 3 or 2 times larger, respectively, than their estimated standard error. All other branches were at least three times larger than their standard deviation. See experimental procedures for further details on the phylogenetic reconstruction methods used. Species whose A-subunit contains an intein are indicated by arrows.
Figure 4
SDS polyacrylamide gel electrophoresis of proteins from induced E. coli. Panel A, Lanes 1 and 3: E. coli Bl21-CodonPlus(DE3)-RIL strain transformed with empty pET-11a vector (negative control); lanes 2, 4, 5 and 6: E. coli Bl21-CodonPlus(DE3)-RIL strain transformed with the Thermoplasma A-ATPase cloned into pET-11a; lane 1-4: cells were induced for 4 hours; lane 5: cells were induced at 16°C for 16 hours; lane 6: cells were induced at 42°C for 2 hours; Sup.: supernatant. Panel B depicts the splicing process schematically. The unprocessed T. acidophilum A-ATPase A subunit has a calculated size of 85 kDalton. The expected molecular weight of the intein is 20 kDalton, and the religated host protein weighs approximately 65 kDalton.
Figure 5
Non-denaturing polyacrylamide gel electrophoresis of proteins from induced E. coli. Lanes 1 and 2: non-denaturing polyacrylamide gel electrophoresis of proteins from the supernatant of the cell lysate. Arrows point towards bands that are present in induced E. coli transformed with the Thermoplasma A-ATPase, but absent in the negative control. The sample buffer did not contain DTT or SDS. Lanes 3, 4, 5 and 6 are separations in denaturing SDS polyacrylamide gels. Lane 3: electro-eluted protein from lane 2 (arrow A); lane 4: electro-eluted protein from lane 2 (arrow B); lanes 1 and 5: E. coli Bl21-CodonPlus(DE3)-RIL strain transformed with empty pET-11a vector (negative control); lanes 2 and 6: E. coli Bl21-CodonPlus(DE3)-RIL strain transformed with the Thermoplasma A-ATPase cloned into pET-11a and induced for 8 hours at 37°C.
Similar articles
- Structure of a putative lipoate protein ligase from Thermoplasma acidophilum and the mechanism of target selection for post-translational modification.
McManus E, Luisi BF, Perham RN. McManus E, et al. J Mol Biol. 2006 Feb 24;356(3):625-37. doi: 10.1016/j.jmb.2005.11.057. Epub 2005 Dec 5. J Mol Biol. 2006. PMID: 16384580 Free PMC article. - Cloning, sequencing and expression of VAT, a CDC48/p97 ATPase homologue from the archaeon Thermoplasma acidophilum.
Pamnani V, Tamura T, Lupas A, Peters J, Cejka Z, Ashraf W, Baumeister W. Pamnani V, et al. FEBS Lett. 1997 Mar 10;404(2-3):263-8. doi: 10.1016/s0014-5793(97)00138-5. FEBS Lett. 1997. PMID: 9119075 - Thermostability and thermoactivity of citrate synthases from the thermophilic and hyperthermophilic archaea, Thermoplasma acidophilum and Pyrococcus furiosus.
Arnott MA, Michael RA, Thompson CR, Hough DW, Danson MJ. Arnott MA, et al. J Mol Biol. 2000 Dec 8;304(4):657-68. doi: 10.1006/jmbi.2000.4240. J Mol Biol. 2000. PMID: 11099387 - The nuclear-encoded inteins of fungi.
Poulter RT, Goodwin TJ, Butler MI. Poulter RT, et al. Fungal Genet Biol. 2007 Mar;44(3):153-79. doi: 10.1016/j.fgb.2006.07.012. Epub 2006 Oct 13. Fungal Genet Biol. 2007. PMID: 17046294 Review. - Protein-splicing intein: Genetic mobility, origin, and evolution.
Liu XQ. Liu XQ. Annu Rev Genet. 2000;34:61-76. doi: 10.1146/annurev.genet.34.1.61. Annu Rev Genet. 2000. PMID: 11092822 Review.
Cited by
- Conservation of intron and intein insertion sites: implications for life histories of parasitic genetic elements.
Swithers KS, Senejani AG, Fournier GP, Gogarten JP. Swithers KS, et al. BMC Evol Biol. 2009 Dec 31;9:303. doi: 10.1186/1471-2148-9-303. BMC Evol Biol. 2009. PMID: 20043855 Free PMC article. - Posttranslational protein modification in Archaea.
Eichler J, Adams MW. Eichler J, et al. Microbiol Mol Biol Rev. 2005 Sep;69(3):393-425. doi: 10.1128/MMBR.69.3.393-425.2005. Microbiol Mol Biol Rev. 2005. PMID: 16148304 Free PMC article. Review. - Exploring the genomic diversity of black yeasts and relatives (Chaetothyriales, Ascomycota).
Teixeira MM, Moreno LF, Stielow BJ, Muszewska A, Hainaut M, Gonzaga L, Abouelleil A, Patané JS, Priest M, Souza R, Young S, Ferreira KS, Zeng Q, da Cunha MM, Gladki A, Barker B, Vicente VA, de Souza EM, Almeida S, Henrissat B, Vasconcelos AT, Deng S, Voglmayr H, Moussa TA, Gorbushina A, Felipe MS, Cuomo CA, de Hoog GS. Teixeira MM, et al. Stud Mycol. 2017 Mar;86:1-28. doi: 10.1016/j.simyco.2017.01.001. Epub 2017 Jan 27. Stud Mycol. 2017. PMID: 28348446 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources