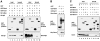Functional cooperation of Dam1, Ipl1, and the inner centromere protein (INCENP)-related protein Sli15 during chromosome segregation - PubMed (original) (raw)
Functional cooperation of Dam1, Ipl1, and the inner centromere protein (INCENP)-related protein Sli15 during chromosome segregation
J Kang et al. J Cell Biol. 2001.
Abstract
We have shown previously that Ipl1 and Sli15 are required for chromosome segregation in Saccharomyces cerevisiae. Sli15 associates directly with the Ipl1 protein kinase and these two proteins colocalize to the mitotic spindle. We show here that Sli15 stimulates the in vitro, and likely in vivo, kinase activity of Ipl1, and Sli15 facilitates the association of Ipl1 with the mitotic spindle. The Ipl1-binding and -stimulating activities of Sli15 both reside within a region containing homology to the metazoan inner centromere protein (INCENP). Ipl1 and Sli15 also bind to Dam1, a microtubule-binding protein required for mitotic spindle integrity and kinetochore function. Sli15 and Dam1 are most likely physiological targets of Ipl1 since Ipl1 can phosphorylate both proteins efficiently in vitro, and the in vivo phosphorylation of both proteins is reduced in ipl1 mutants. Some dam1 mutations exacerbate the phenotype of ipl1 and sli15 mutants, thus providing evidence that Dam1 interactions with Ipl1-Sli15 are functionally important in vivo. Similar to Dam1, Ipl1 and Sli15 each bind to microtubules directly in vitro, and they are associated with yeast centromeric DNA in vivo. Given their dual association with microtubules and kinetochores, Ipl1, Sli15, and Dam1 may play crucial roles in regulating chromosome-spindle interactions or in the movement of kinetochores along microtubules.
Figures
Figure 1.
Properties of different regions of Sli15. The region of putative coiled-coil is filled in. The putative nuclear localization signal is shown as a white dot. The conserved IN box is stippled.
Figure 2.
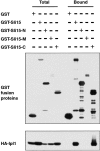
The IN box–containing COOH-terminal region of Sli15 binds Ipl1. GST (pEG[KT]), GST–Sli15 (pCC1061), GST–Sli15-N (pCC1563), GST–Sli15-M (pCC1564), or GST–Sli15-C (pCC1565) was affinity-purified with glutathione-agarose beads from a yeast strain (TD4) that also expressed HA–Ipl1 (pCC1128). Aliquots of proteins from total lysates (Total) or from the affinity-purified (Bound) fractions (after three rounds of washing with a buffer containing 200 mM KCl) were analyzed by immunoblotting using antibodies against GST or the HA epitope. The amount of bound fraction loaded was 4.5-fold that of the total lysates.
Figure 3.
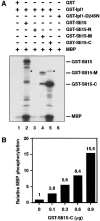
Ipl1 phosphorylates Sli15 and is activated by Sli15. (A) In vitro phosphorylation of Sli15 by Ipl1 and [γ-32P]ATP. With the exception of bovine MBP, all proteins used were purified from E. coli. The asterisks denote labeled GST–Ipl1 bands caused by autophosphorylation. (B) Stimulation of GST–Ipl1 kinase activity by GST–Sli15-C. MBP (1 μg) was phosphorylated by GST–Ipl1 (0.6 μg) in the presence of various amounts of GST–Sli15-C. (C) Phosphorylation of Sli15 is altered in ipl1 mutant cells. Wild-type (WT) (CCY766-9D) or ipl1-2 (CCY915-13C) cells carrying pCC1301 (Sli15–HA) were incubated in supplemented synthetic complete (SC) medium lacking leucine at 26°C and then shifted to 37°C for 2.5 h. Protein extracts from these cells were analyzed by immunoblotting with anti-HA or anti-G6PDH antibodies.
Figure 3.
Ipl1 phosphorylates Sli15 and is activated by Sli15. (A) In vitro phosphorylation of Sli15 by Ipl1 and [γ-32P]ATP. With the exception of bovine MBP, all proteins used were purified from E. coli. The asterisks denote labeled GST–Ipl1 bands caused by autophosphorylation. (B) Stimulation of GST–Ipl1 kinase activity by GST–Sli15-C. MBP (1 μg) was phosphorylated by GST–Ipl1 (0.6 μg) in the presence of various amounts of GST–Sli15-C. (C) Phosphorylation of Sli15 is altered in ipl1 mutant cells. Wild-type (WT) (CCY766-9D) or ipl1-2 (CCY915-13C) cells carrying pCC1301 (Sli15–HA) were incubated in supplemented synthetic complete (SC) medium lacking leucine at 26°C and then shifted to 37°C for 2.5 h. Protein extracts from these cells were analyzed by immunoblotting with anti-HA or anti-G6PDH antibodies.
Figure 4.
GST–Ipl1 and GST–Sli15-M cosediment with microtubules. Different concentrations of taxol-stabilized microtubules were incubated with GST (A), GST–Ipl1 (B), or GST–Sli15-M (C). The reactions were then centrifuged at high speed to generate the supernatant (S) and pellet (P) fractions. The presence of GST or GST fusion protein in either fraction was determined by immunoblotting with anti-GST antibodies. The percentage of GST–Ipl1 or GST–Sli15-M that was bound to and copelleted with the microtubules in each reaction was quantitated by densitometry of the immunoblots. The binding curves display the average and range of values obtained from duplicate experiments.
Figure 5.
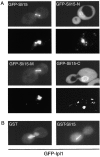
Subcellular localization of GFP–Sli15 and GFP–Ipl1. (A) Subcellular localization of full-length and truncated forms of Sli15. Wild-type diploid yeast cells (DBY1830) expressing GFP–Sli15 (pCC1060), GFP–Sli15-N (pCC1533), GFP–Sli15-M (pCC1534), or GFP–Sli15-C (pCC1566) were stained with DAPI and examined. For each pair of images: top, GFP fusion protein image; bottom, DAPI-stained image. (B) Subcellular localization of GFP–Ipl1. Wild-type haploid yeast cells (TD4) carrying pCC1584 (GFP–Ipl1) and pEG(KT) (GST) or carrying pCC1584 and pCC1061 (GST–Sli15) were cultured for 2 h in medium containing 4% galactose to induce expression of GST or GST–Sli15, respectively. Images of GFP–Ipl1 are shown.
Figure 6.
Dam1 binds to Ipl1 and Sli15. (A) In vivo association of Ipl1 or Sli15 with Dam1 or Duo1. GST (pEG[KT]), GST–Dam1 (pDD1017), or GST–Duo1 (pDD475) were affinity-purified with glutathione-agarose beads from a yeast strain (TD4) that also expressed HA–Ipl1 (pCC1128) or Sli15–Myc (pCC1173). Aliquots of proteins from total lysates (total) or from the affinity-purified (bound) fractions (after three rounds of washing with a buffer containing 200 mM KCl) were analyzed by immunoblotting using anti-GST, anti-HA, or anti-Myc antibodies. (B) Direct binding of Dam1 to Ipl1 and Sli15. GST (pGEX-2T), GST–Sli15 (pCC1062), and GST–Ipl1 (pCC669) were affinity-purified with glutathione-agarose beads from E. coli and tested for their ability to bind His6–Dam1 (pDD884). Proteins were detected by immunoblotting with anti-GST or anti-Dam1 antibodies. (C) The middle and COOH-terminal regions of Sli15 bind Dam1. The ability of GST (pEG[KT]), GST–Sli15 (pCC1061), GST–Sli15-N (pCC1563), GST–Sli15-M (pCC1564), or GST–Sli15-C (pCC1565) to associate with Dam1 (pCC1575) was determined as in A. The amount of bound fraction loaded in A and C was 4.5-fold that of the total lysates.
Figure 7.
Ipl1 phosphorylates Dam1. (A) In vitro phosphorylation of Dam1 by Ipl1 and [γ-32P]ATP. With the exception of bovine MBP and GST–Duo1 (from yeast), all proteins used were purified from E. coli. (B) Dam1 is phosphorylated in vivo. Dam1, immunoprecipitated from yeast cells (DDY904), was treated with λ phosphatase and analyzed by immunoblotting with anti-Dam1 antibodies. (C) Phosphorylation of Dam1 is altered in ipl1 and sli15 mutant cells. Wild-type (WT) (DDY904), ipl1-2 (CCY915-13C), or sli15-3 (CCY1124-4B) cells were incubated in YEPD medium at 25°C and then shifted to 37°C for 3 h. Protein extracts from these cells were analyzed by immunoblotting with anti-Dam1 antibodies.
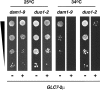
Figure 8.
Exacerbation of temperature sensitivity of dam1 and duo1 mutants by increased dosage of GLC7. Suspensions of dam1-9 (DDY1906) and duo1-2 (DDY1525) cells carrying the control plasmid pRS316 (−) or the _GLC7_-2μ plasmid pCC510 (+) were diluted 10-fold serially, spotted on SC medium lacking uracil, and allowed to grow at the indicated temperatures.
Figure 9.
Association of Ipl1 and Sli15 with centromeric DNA. Extracts prepared from formaldehyde-fixed yeast cells (CCY776-5D) that did not (untagged) or did express HA–Ipl1 (pCC1128) or Sli15–Myc (pCC1193) were prepared. These extracts were left untreated (WCE), mock-precipitated without antibodies (No Ab), or immunoprecipitated with anti-HA (α HA) or anti-Myc (α Myc) antibodies. PCR was performed to amplify DNA fragments from WCE or precipitated samples, using primers that flank CEN3 or CEN16, or primers from a region located ∼1 kb from CEN16.
Similar articles
- Ipl1/Aurora-dependent phosphorylation of Sli15/INCENP regulates CPC-spindle interaction to ensure proper microtubule dynamics.
Nakajima Y, Cormier A, Tyers RG, Pigula A, Peng Y, Drubin DG, Barnes G. Nakajima Y, et al. J Cell Biol. 2011 Jul 11;194(1):137-53. doi: 10.1083/jcb.201009137. Epub 2011 Jul 4. J Cell Biol. 2011. PMID: 21727193 Free PMC article. - Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14.
Pereira G, Schiebel E. Pereira G, et al. Science. 2003 Dec 19;302(5653):2120-4. doi: 10.1126/science.1091936. Epub 2003 Nov 6. Science. 2003. PMID: 14605209 - A Bir1-Sli15 complex connects centromeres to microtubules and is required to sense kinetochore tension.
Sandall S, Severin F, McLeod IX, Yates JR 3rd, Oegema K, Hyman A, Desai A. Sandall S, et al. Cell. 2006 Dec 15;127(6):1179-91. doi: 10.1016/j.cell.2006.09.049. Cell. 2006. PMID: 17174893 Free PMC article. - Protein kinases in mitotic phosphorylation of budding yeast CENP-A.
Mishra PK, Basrai MA. Mishra PK, et al. Curr Genet. 2019 Dec;65(6):1325-1332. doi: 10.1007/s00294-019-00997-5. Epub 2019 May 22. Curr Genet. 2019. PMID: 31119371 Review. - Structures and functions of yeast kinetochore complexes.
Westermann S, Drubin DG, Barnes G. Westermann S, et al. Annu Rev Biochem. 2007;76:563-91. doi: 10.1146/annurev.biochem.76.052705.160607. Annu Rev Biochem. 2007. PMID: 17362199 Review.
Cited by
- Deficient sumoylation of yeast 2-micron plasmid proteins Rep1 and Rep2 associated with their loss from the plasmid-partitioning locus and impaired plasmid inheritance.
Pinder JB, McQuaid ME, Dobson MJ. Pinder JB, et al. PLoS One. 2013;8(3):e60384. doi: 10.1371/journal.pone.0060384. Epub 2013 Mar 28. PLoS One. 2013. PMID: 23555963 Free PMC article. - Cleavage furrow positioning.
Glotzer M. Glotzer M. J Cell Biol. 2004 Feb 2;164(3):347-51. doi: 10.1083/jcb.200310112. J Cell Biol. 2004. PMID: 14757750 Free PMC article. Review. - Chromosomal enrichment and activation of the aurora B pathway are coupled to spatially regulate spindle assembly.
Kelly AE, Sampath SC, Maniar TA, Woo EM, Chait BT, Funabiki H. Kelly AE, et al. Dev Cell. 2007 Jan;12(1):31-43. doi: 10.1016/j.devcel.2006.11.001. Dev Cell. 2007. PMID: 17199039 Free PMC article. - Global control of histone modification by the anaphase-promoting complex.
Ramaswamy V, Williams JS, Robinson KM, Sopko RL, Schultz MC. Ramaswamy V, et al. Mol Cell Biol. 2003 Dec;23(24):9136-49. doi: 10.1128/MCB.23.24.9136-9149.2003. Mol Cell Biol. 2003. PMID: 14645525 Free PMC article. - Aurora B hyperactivation by Bub1 overexpression promotes chromosome missegregation.
Ricke RM, van Deursen JM. Ricke RM, et al. Cell Cycle. 2011 Nov 1;10(21):3645-51. doi: 10.4161/cc.10.21.18156. Epub 2011 Nov 1. Cell Cycle. 2011. PMID: 22033440 Free PMC article. Review.
References
- Adams, R.R., S.P. Wheatley, A.M. Gouldsworthy, S.E. Kandels-Lewis, M. Carmena, C. Smythe, D.L. Gerloff, and W.C. Earnshaw. 2000. INCENP binds the Aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr. Biol. 10:1075–1078. - PubMed
- Bischoff, F.R., and G.D. Plowman. 1999. The Aurora/Ipl1p kinase family: regulators of chromosome segregation and cytokinesis. Trends Cell Biol. 9:454–459. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases