Laribacter hongkongensis gen. nov., sp. nov., a novel gram-negative bacterium isolated from a cirrhotic patient with bacteremia and empyema - PubMed (original) (raw)
Case Reports
Laribacter hongkongensis gen. nov., sp. nov., a novel gram-negative bacterium isolated from a cirrhotic patient with bacteremia and empyema
K Y Yuen et al. J Clin Microbiol. 2001 Dec.
Abstract
A bacterium was isolated from the blood and empyema of a cirrhotic patient. The cells were facultatively anaerobic, nonsporulating, gram-negative, seagull shaped or spiral rods. The bacterium grows on sheep blood agar as nonhemolytic, gray colonies 1 mm in diameter after 24 h of incubation at 37 degrees C in ambient air. Growth also occurs on MacConkey agar and at 25 and 42 degrees C but not at 4, 44, and 50 degrees C. The bacterium can grow in 1 or 2% but not 3, 4, or 5% NaCl. No enhancement of growth is observed with 5% CO(2). The organism is aflagellated and nonmotile at both 25 and 37 degrees C. It is oxidase, catalase, urease, and arginine dihydrolase positive, and it reduces nitrate. It does not ferment, oxidize, or assimilate any sugar tested. 16S rRNA gene sequencing showed that there are 91 base differences (6.2%), 112 base differences (7.7%), and 116 base differences (8.2%) between the bacterium and Microvirgula aerodenitrificans, Vogesella indigofera, and Chromobacterium species, respectively. The G+C content (mean and standard deviation) is 68.0% +/- 2.43%, and the genomic size is about 3 Mb. Based on phylogenetic affiliation, the bacterium belongs to the Neisseriaceae family of the beta-subclass of Proteobacteria. For these reasons, a new genus and species, Laribacter hongkongensis gen. nov., sp. nov., is proposed, for which HKU1 is the type strain. Further studies should be performed to ascertain the potential of this bacterium to become an emerging pathogen.
Figures
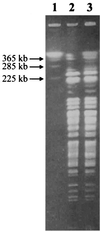
FIG. 1
Pulsed-field gel electrophoresis of genomic DNA of HKU1 recovered from blood (lane 2) and empyema (lane 3) after _Xba_I digestion and S. cerevisiae chromosomal DNA size marker (lane 1).
FIG. 2
Scanning electron micrograph of L. hongkongensis. Cells vary in length from 0.79 to 2.5 μm. The bacterium has a spiral curvature and is aflagellated. Bar, 1 μm.
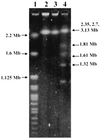
FIG. 3
Pulsed-field gel electrophoresis of genomic DNA of HKU1 recovered from blood (lane 2) and empyema (lane 3), H. wingel chromosomal DNA size marker (lane 1), and S. cerevisiae chromosomal DNA size marker (lane 4), showing that the genome size of HKU1 is about 3 Mb.
FIG. 4
Phylogenetic tree showing the relationship of L. hongkongensis gen. nov., sp. nov., to the other members of the β-subclass of Proteobacteria. A total of 1,398 nucleotide positions in each 16S rRNA gene were included in the analysis. The scale bar indicates the estimated number of substitutions per 100 bases using the Jukes-Cantor correction; the length of the bar represents 1 substitution. Names and accession numbers are given as cited in the GenBank database.
References
- Ausubel F M, Brent R, Kingston R E, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1998. pp. 2.4.1–2.4.2.
- Bauer A W, Kirby W M M, Sherris J C, Turck M. Antibiotic susceptibility testing by a single disc method. Am J Clin Pathol. 1966;45:493. - PubMed
- Cheuk W, Woo P C Y, Yuen K Y, Yu P H, Chan J K C. Intestinal inflammatory pseudotumor with regional lymph node involvment: identification of a new bacterium as the etiologic agent. J Pathol. 2001;192:289–292. - PubMed
- Goodfellow M. Chemical methods in bacterial systematics. London, England: Academic Press Ltd.; 1985. pp. 67–93.
- Luk W K, Wong S S, Yuen K Y, Ho P L, Woo P C Y, Lee R A, Chau P Y. Inpatient emergencies encountered by an infectious disease consultative service. Clin Infect Dis. 1998;26:695–701. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous