Targeting of human DNA polymerase iota to the replication machinery via interaction with PCNA - PubMed (original) (raw)
Targeting of human DNA polymerase iota to the replication machinery via interaction with PCNA
L Haracska et al. Proc Natl Acad Sci U S A. 2001.
Abstract
Human DNA polymerase iota (hPoliota) promotes translesion synthesis by inserting nucleotides opposite highly distorting or noninstructional DNA lesions. Here, we provide evidence for the physical interaction of hPoliota with proliferating cell nuclear antigen (PCNA), and show that PCNA, together with replication factor C (RFC) and replication protein A (RPA), stimulates the DNA synthetic activity of hPoliota. In the presence of these protein factors, on undamaged DNA, the efficiency (V(max)/K(m)) of correct nucleotide incorporation by hPoliota is increased approximately 80-150-fold, and this increase in efficiency results from a reduction in the apparent K(m) for the nucleotide. PCNA, RFC, and RPA also stimulate nucleotide incorporation opposite the 3'-T of the (6) thymine-thymine (T-T) photoproduct and opposite an abasic site. The interaction of hPoliota with PCNA implies that the targeting of this polymerase to the replication machinery stalled at a lesion site is achieved via this association.
Figures
Figure 1
Interaction of hPolι with PCNA. hPolι was incubated with human PCNA and gel-filtered (Superdex 200). (I) PCNA is alone (5 μg); (II) the mixture of hPolι (4 μg) and PCNA (5 μg) were gel filtered. Column fractions (Fr.) are indicated, and the positions of molecular weight standards are shown on the left. The elution positions of molecular weight markers for the gel filtration column are indicated on top. PCNA and hPolι are identified on the right.
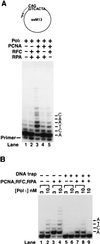
Figure 2
Effect of PCNA, RFC, and RPA on hPolι activity. (A) DNA synthesis by hPolι in the presence of different combinations of PCNA, RFC, and RPA. The complete standard reaction mixture contained hPolι (10 nM), PCNA (100 ng), RFC (50 ng), and RPA (250 ng), and all four deoxynucleotides (100 μM each) along with a ssM13 DNA (2 nM) primed with a 5′ 32P-labeled 38-nt oligonucleotide. Various combinations of PCNA, RFC, and RPA were added to the reaction mixture as indicated. (B) Processivity of hPolι in the presence of PCNA, RFC, and RPA. hPolι (3 nM or 10 nM) alone (lanes 1 and 2, and 5 and 6), or in the presence of PCNA (100 ng), RFC (50 ng), and RPA (250 ng; lanes 3 and 4, and 7 and 8), was preincubated with circular ssM13 template DNA (10 nM) singly primed with a 5′ 32P-labeled oligonucleotide for 5 min at 37°C. Primer extension reactions were initiated by adding all four deoxynucleotides (500 μM each; lanes 1–4) or all four deoxynucleotides and excess sonicated herring sperm DNA (0.5 mg/ml) as a trap (lanes 5–8). After incubation for 10 min at 37°C, samples were quenched and run on a 10% polyacrylamide gel. To demonstrate the effectiveness of the trap, hPolι, along with PCNA, RFC, and RPA, was preincubated with the trap DNA together with the DNA substrate before addition of dNTPs (lane 9).
Figure 3
Stimulation of nucleotide incorporation by hPolι in the presence of PCNA, RFC, and RPA on lesion containing DNA substrates. (A) Effect of PCNA, RFC, and RPA on DNA synthesis on an undamaged linear primer:template substrate bearing biotin–streptavidin complex at the ends. The complete reaction mixture contained hPolι (3 nM), PCNA (100 ng), RFC (50 ng), RPA (50 ng), and all four deoxynucleotides (100 μM each) along with the DNA substrate (20 nM). Various combinations of PCNA, RFC, and RPA were added to the reaction mixture as indicated. (B) DNA synthesis by hPolι on lesion containing DNA substrates in the presence or absence of PCNA, RFC, and RPA. A portion of the running start DNA substrate containing the undamaged T-T residues, a cis_-syn T-T dimer, or a (–4) T-T photoproduct is shown in I. The position of the undamaged or damaged TT residues is indicated by asterisks. In the abasic site containing DNA substrate shown in_II, the abasic site in the template DNA is indicated by0 and marked by an asterisk. Polι (3 nM) was incubated with the DNA substrate (20 nM) in the presence of the four dNTPs (100 μM each) under standard reaction conditions.
Figure 4
Deoxynucleotide incorporation opposite undamaged G or T residues, and opposite a _cis_-syn T-T dimer, a (–4) T-T photoproduct, and an abasic (AP) site by hPolι. The linear DNA substrates contained the biotin–streptavidin complex on both ends. hPolι (1 nM) was incubated with primer:template DNA substrate (20 nM) and increasing concentrations of a single deoxynucleotide in the absence or presence of PCNA (100 ng), RFC (50 ng), and RPA (50 ng).
Similar articles
- Stimulation of DNA synthesis activity of human DNA polymerase kappa by PCNA.
Haracska L, Unk I, Johnson RE, Phillips BB, Hurwitz J, Prakash L, Prakash S. Haracska L, et al. Mol Cell Biol. 2002 Feb;22(3):784-91. doi: 10.1128/MCB.22.3.784-791.2002. Mol Cell Biol. 2002. PMID: 11784855 Free PMC article. - A single domain in human DNA polymerase iota mediates interaction with PCNA: implications for translesion DNA synthesis.
Haracska L, Acharya N, Unk I, Johnson RE, Hurwitz J, Prakash L, Prakash S. Haracska L, et al. Mol Cell Biol. 2005 Feb;25(3):1183-90. doi: 10.1128/MCB.25.3.1183-1190.2005. Mol Cell Biol. 2005. PMID: 15657443 Free PMC article. - Interaction with PCNA is essential for yeast DNA polymerase eta function.
Haracska L, Kondratick CM, Unk I, Prakash S, Prakash L. Haracska L, et al. Mol Cell. 2001 Aug;8(2):407-15. doi: 10.1016/s1097-2765(01)00319-7. Mol Cell. 2001. PMID: 11545742 - 'PIPs' in DNA polymerase: PCNA interaction affairs.
Acharya N, Patel SK, Sahu SR, Kumari P. Acharya N, et al. Biochem Soc Trans. 2020 Dec 18;48(6):2811-2822. doi: 10.1042/BST20200678. Biochem Soc Trans. 2020. PMID: 33196097 Review. - Functions of Multiple Clamp and Clamp-Loader Complexes in Eukaryotic DNA Replication.
Ohashi E, Tsurimoto T. Ohashi E, et al. Adv Exp Med Biol. 2017;1042:135-162. doi: 10.1007/978-981-10-6955-0_7. Adv Exp Med Biol. 2017. PMID: 29357057 Review.
Cited by
- Separate roles of structured and unstructured regions of Y-family DNA polymerases.
Ohmori H, Hanafusa T, Ohashi E, Vaziri C. Ohmori H, et al. Adv Protein Chem Struct Biol. 2009;78:99-146. doi: 10.1016/S1876-1623(08)78004-0. Epub 2009 Nov 27. Adv Protein Chem Struct Biol. 2009. PMID: 20663485 Free PMC article. Review. - Human DNA polymerase iota promotes replication through a ring-closed minor-groove adduct that adopts a syn conformation in DNA.
Wolfle WT, Johnson RE, Minko IG, Lloyd RS, Prakash S, Prakash L. Wolfle WT, et al. Mol Cell Biol. 2005 Oct;25(19):8748-54. doi: 10.1128/MCB.25.19.8748-8754.2005. Mol Cell Biol. 2005. PMID: 16166652 Free PMC article. - Regulation of Saccharomyces cerevisiae DNA polymerase eta transcript and protein.
Pabla R, Rozario D, Siede W. Pabla R, et al. Radiat Environ Biophys. 2008 Feb;47(1):157-68. doi: 10.1007/s00411-007-0132-1. Epub 2007 Sep 14. Radiat Environ Biophys. 2008. PMID: 17874115 - Genetic control of predominantly error-free replication through an acrolein-derived minor-groove DNA adduct.
Yoon JH, Hodge RP, Hackfeld LC, Park J, Roy Choudhury J, Prakash S, Prakash L. Yoon JH, et al. J Biol Chem. 2018 Feb 23;293(8):2949-2958. doi: 10.1074/jbc.RA117.000962. Epub 2018 Jan 12. J Biol Chem. 2018. PMID: 29330301 Free PMC article. - Translesion Synthesis: Insights into the Selection and Switching of DNA Polymerases.
Zhao L, Washington MT. Zhao L, et al. Genes (Basel). 2017 Jan 10;8(1):24. doi: 10.3390/genes8010024. Genes (Basel). 2017. PMID: 28075396 Free PMC article. Review.
References
- Goodman M F, Tippin B. Curr Opin Genet Dev. 2000;10:162–168. - PubMed
- Johnson R E, Prakash S, Prakash L. Science. 1999;283:1001–1004. - PubMed
- Johnson R E, Washington M T, Prakash S, Prakash L. J Biol Chem. 2000;275:7447–7450. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous