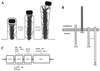Actin-based motility of intracellular microbial pathogens - PubMed (original) (raw)
Review
Actin-based motility of intracellular microbial pathogens
M B Goldberg. Microbiol Mol Biol Rev. 2001 Dec.
Abstract
A diverse group of intracellular microorganisms, including Listeria monocytogenes, Shigella spp., Rickettsia spp., and vaccinia virus, utilize actin-based motility to move within and spread between mammalian host cells. These organisms have in common a pathogenic life cycle that involves a stage within the cytoplasm of mammalian host cells. Within the cytoplasm of host cells, these organisms activate components of the cellular actin assembly machinery to induce the formation of actin tails on the microbial surface. The assembly of these actin tails provides force that propels the organisms through the cell cytoplasm to the cell periphery or into adjacent cells. Each of these organisms utilizes preexisting mammalian pathways of actin rearrangement to induce its own actin-based motility. Particularly remarkable is that while all of these microbes use the same or overlapping pathways, each intercepts the pathway at a different step. In addition, the microbial molecules involved are each distinctly different from the others. Taken together, these observations suggest that each of these microbes separately and convergently evolved a mechanism to utilize the cellular actin assembly machinery. The current understanding of the molecular mechanisms of microbial actin-based motility is the subject of this review.
Figures
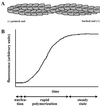
FIG. 1
Pathogenesis of Shigella (representative of the pathogenesis of Listeria and Rickettsia as well). 1, Shigella organisms (solid ellipses) enter mammalian host cells by inducing phagocytosis. 2 to 4, After entry, the bacterium is within a phagocytic vacuole (step 2), which it lyses (step 3), thereby releasing it into the cytoplasm of the host cell (step 4). 5, the bacterium assembles an actin tail on one pole. Assembly of the actin tail propels it through the cell cytoplasm. 6, Actin tail assembly also enables it to form a protrusion from the cell surface. The protrusion contacts the membrane of the adjacent cell and is taken up, along with the bacterium within it. 7 to 9, The bacterium is then within a double-membrane vacuole, which it lyses, thereby releasing it into the cytoplasm of the adjacent cell. 10, The bacterium again assembles an actin tail that propels it through the cell.
FIG. 2
Actin filament structure and assembly dynamics. (A) Asymmetric homopolymer of helically arranged actin. The pointed or minus end (left) and barbed or plus end (right) are indicated. ATP bound to the actin monomer is hydrolyzed to ADP shortly after addition to the filament. (B) Actin polymerization curve. An idealized curve from a pyrene actin assay is shown. Actin is polymerized in vitro under experimental test conditions. At time zero, potassium and magnesium are added. The lag phase represents the time required for actin nucleation. The rapid polymerization phase represents the time during which short filaments elongate. Steady state represents an equilibrium between growth of the filaments due to monomer addition and shortening of the filaments due to loss of monomer.
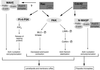
FIG. 3
Signaling pathways involving the small GTPases Cdc42 and Rac. Shown is an overview of the Cdc42 and Rac signal transduction pathways that lead to actin rearrangements in the form of lamellipodia (membrane ruffles) and filopodia (microspikes). (Adapted from reference with permission of the publisher.)
FIG. 4
Dendritic network at the leading edge of a lamellopodium of a motile cell. (A) Branching of actin filaments at approximately 70° angles, consistent with Arp2/3-mediated cross-linking. (B and C) Higher magnification of boxes b and c, respectively, in panel A. (D) Dendritic nucleation model. The proposed model of Arp2/3 complex-mediated nucleation and branching of actin filaments is shown. The Arp2/3 complex binds to the side of an actin filament and is bound by a WASP family member (in this diagram, Scar). Both binding to the side of the filament and binding by a WASP family member are thought to play a role in activation of Arp2/3 complex nucleation of the new filament on the side of the mother filament. (E) Proposed model for the formation of branched networks of actin filaments in lamellipodia at the leading edge of motile cells. The Arp2/3 complex, in conjunction with a WASP family member (not shown), nucleates actin filaments on the sides of existing filaments. Cofilin severs filaments to generate additional uncapped barbed ends that can polymerize quickly. These processes lead to the rapid extension of an actin filament network that generates the force to push the cell membrane forward. (Panels A, B, C, and E reprinted from reference with permission of the publisher. Panel D reprinted from reference with permission of the publisher.
FIG. 5
Domain structure of WASP family members. Black, WH1 domain; blue, GBD; yellow, proline-rich region (number indicates the number of motifs containing a stretch of five or more prolines); green, WH2 or verprolin homology domain; pink, acidic domain (numbers indicate the numbers of acidic/basic residues). The cofilin homology domain lies between the WH2 and acidic domains and is not specifically indicated. Numbers to the right indicate length in amino acids. (Adapted from reference with permission of the publisher.)
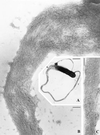
FIG. 6
Thin-section electron microscopy of an actin tail formed by an intracellular L. monocytogenes bacterium. Note that the tail consists of short actin filaments that are bundled in a nonparallel fashion. (Reprinted from reference with permission of the publisher.)
FIG. 7
Negative-staining electron microscopy of an actin tail formed by an L. monocytogenes bacterium within a cell surface protrusion. Note that the tail consists of long actin filaments that are bundled in a parallel fashion. (Reprinted from reference with permission of the publisher)
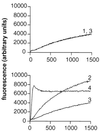
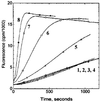
FIG. 8
ActA stimulation of Arp2/3 complex-mediated actin polymerization. Fluorescence intensity is plotted as a function of time in pyrene-actin polymerization assays. 1, actin; 2, actin and Arp2/3 complex; 3, actin and ActA (amino acids 29 to 263); 4, actin, Arp2/3 complex, and ActA (amino acids 29 to 263). (Reprinted from reference with permission of the publisher.)
FIG. 9
Functional domains of L. monocytogenes ActA. (A) Schematic of ActA. SP, signal peptide; A, WASP-like acidic domain, possibly involved in Arp2/3 complex binding; WH2, WASP homology 2 domain; PI, phosphoinositide binding region; Pro-rich repeats, proline-rich repeats; TM, transmembrane anchor; AB, monomeric actin-binding region, amino acids 60 to 100; dimer, region required for ActA dimerization, amino acids 126 to 155; clouds/tails, region important for formation of actin tails, rather than actin clouds, amino acids 166 to 256. (B) Alignment of ActA amino acids 85 to 99 with the first of two WH2 domains of N-WASP and of ActA amino acids 121 to 170 with the WH2 and Arp2/3 complex-binding domains of WASP family members. (C) Alignment of ActA amino acids 32 to 45 with the acidic domains of WASP family members. Sequences used for alignment: mouse WASP, human N-WASP, and human Scar1. (Panel B adapted from reference with permission of the publisher. Panel C adapted from reference with permission of the publisher.)
FIG. 10
Proposed location of the ActA-binding site on the Arp2/3 complex and on the Arp2 subunit. (A) Proposed site for binding of ActA, Scar, and N-WASP to the nearest-neighbor model of the Arp2/3 complex. Shading indicates subunits to which ActA, Scar, and N-WASP cross-link. Open circles and dark lines indicate subunits that cross-link within the complex (118). Ovals indicate interactions demonstrated by two-hybrid analysis (95). (B) Profilin- and actophorin (cofilin)-binding sites on conventional actin (13, 53, 154). (C) Proposed structure of the Arp2 subunit, based on homology to conventional actin (73). The binding site of ActA that has been proposed on the basis of competition experiments (see the text) is indicated. (Reprinted from reference with permission of the publisher.)
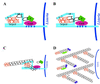
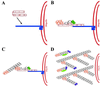
FIG. 11
Model of actin tail assembly by Listeria ActA. (A) ActA (light blue bar) binds the Arp2/3 complex (orange) at its Arp2/3 complex-binding domain (amino acids 143 to 169, designated Arp2/3) and possibly at its acidic domain (amino acids 31 to 58, designated A). ActA binds an actin monomer (black hexagon) at its actin-binding domain (amino acids 60 to 100, designated AB). ActA binds VASP (pink oval) at its proline-rich repeat region (amino acids 293 to 390, designated by four bands), and VASP binds profilin (P, green rectangle)-ATP-actin. (B) The actin nucleation activity of the Arp2/3 complex is stimulated by its binding to ActA. It mediates the addition of actin monomers to the barbed end and caps the pointed end of a new actin filament. (C) As the filament extends, the original Arp2/3 complex is released from ActA and another Arp2/3 complex binds both ActA and the side of an existing actin filament. Each interaction stimulates the actin-nucleating activity of the Arp2/3 complex. (D) Repeated rounds of filament branching, filament nucleation, filament extension, and Arp2/3 complex release generate a network of actin filaments linked at 70° angles. At a distance from the bacterial surface, filament barbed ends are capped, thereby halting the addition of more actin monomers. Also at a distance from the bacterial surface, filaments debranch and, with the assistance of the actin-severing protein cofilin, depolymerize, thereby maintaining a local pool of actin monomers. Whether cofilin also severs filaments near the bacterial surface, thereby generating uncapped barbed ends, is unknown.
FIG. 12
Actin tail formation by IcsA-expressing E. coli. Time-lapse microscopy of IcsA-expressing E. coli in X. laevis oocyte cytoplasmic extracts is shown. IcsA is sufficient to enable E. coli to assemble actin tails. Bar, 10 μm. (Reprinted from reference with permission of the publisher.)
FIG. 13
Actin tail formation by IcsA-coated particles. Fluorescence (top) and phase (bottom) microscopy of IcsA-coated irregularly shaped silica particles in X. laevis oocyte cytoplasmic extracts are shown. Rhodamine actin, which fluoresces on polymerization, has been added to extracts. A single particle breaks away from the clump of particles as it assembles an actin tail. The clump of particles polymerizes actin on its surface (Magdalena and Goldberg, unpublished).
FIG. 14
Domains of IcsA. α domain (amino acids 53 to 758), portion of mature IcsA that is exposed on the bacterial surface; β domain (amino acids 759 to 1102), outer membrane anchor; SP (amino acids 1 to 52), signal peptide; actin assembly, region that is sufficient to polymerize actin in in vitro assays (see the text).
FIG. 15
Unipolar IcsA on the surface of Shigella. Indirect-immunofluorescence (left) and phase (right) microscopy of S. flexneri are shown. Arrows indicate unipolar IcsA on nondividing bacteria; arrowheads indicate bipolar IcsA on a dividing bacterium.
FIG. 16
Fluorescence microscopy showing that actin assembly occurs at the Shigella pole on which IcsA is most highly expressed. IcsA (red) and polymerized actin (green).
FIG. 17
Fluorescence microscopy of _Shigella_-infected cells showing localization of N-WASP to the Shigella pole. (A) Actin (phalloidin); (B) N-WASP antibody; (C) IcsA antibody; (D) overlay of actin and N-WASP antibody, where yellow indicates colocalization. (Reprinted from reference with permission of the publisher.)
FIG. 18
Fluorescence microscopy of _Shigella_-infected cells showing localization of the Arp2/3 complex to actin tails formed by Shigella. (A) Arp3 antibody; (B) actin (phalloidin); (C) p34 antibody; (D) actin (phalloidin).
FIG. 19
IcsA stimulates N-WASP activation of Arp2/3 complex-mediated actin polymerization. Fluorescence intensity is plotted as a function of time in pyrene-actin polymerization assays. 1, actin alone; 2, actin and Arp2/3 complex; 3, actin and 0.25 uM IcsA; 4, actin and N-WASP; 5, actin, Arp2/3 complex, and 0.25 μM IcsA; 6, actin, Arp2/3 complex, and N-WASP; 7, actin, Arp2/3 complex, N-WASP, and 30 nM IcsA; 8, actin, Arp2/3 complex, N-WASP, and 0.25 μM IcsA. (Reprinted from reference with permission of the publisher.)
FIG. 20
Model of actin tail assembly by Shigella IcsA. (A) IcsA (blue bar) on the surface of Shigella binds N-WASP (maroon). (B) IcsA binding disrupts the intramolecular bonds and activates N-WASP, thereby stimulating N-WASP activation of the Arp2/3 complex (orange), N-WASP binding to monomeric actin (black hexagon), and possibly N-WASP binding to profilin-actin (P, green rectangle). The Arp2/3 complex nucleates actin, mediates the addition of actin monomers to the barbed end, and caps the pointed end of a new actin filament. (C) As the filament extends, the original Arp2/3 complex is released from N-WASP and another Arp2/3 complex binds both N-WASP and the side of an existing actin filament. Each interaction stimulates the actin-nucleating activity of the Arp2/3 complex. (D) Repeated rounds of filament branching, filament nucleation, filament extension, and Arp2/3 complex release generate a network of actin filaments linked at 70° angles. At a distance from the bacterial surface, filament barbed ends are capped, thereby halting the addition of more actin monomers. Also at a distance from the bacterial surface, filaments debranch and, with the assistance of the actin-severing protein cofilin, depolymerize, thereby maintaining a local pool of actin monomers. Whether cofilin also severs filaments near the bacterial surface, thereby generating uncapped barbed ends, is unknown. V, verprolin homology domain; C, cofilin homology domain; A, acidic domain; Pro, proline-rich region; GBD, GTPase-binding domain; PH, pleckstrin homology domain.
FIG. 21
Fluorescence microscopy of vaccinia virus-infected HeLa cells, labeled for actin (red) and vaccinia virus IMV particles (green), showing actin tail formation by vaccinia virus. (A) Actin tails on intracellular vaccinia virus. Bar, 50 μm. (B) Protrusions from the cell surface, formed by vaccinia virus, that contain actin tails and have viral particles at the tips. Bar, 10 μm. (Adapted from reference with permission of the publisher.)
FIG. 22
Electron microscopy of filaments within the actin tail that have been decorated with the S1 subfragment of myosin show actin tail formation by vaccinia virus. Stars, viral particles. Bar, 400 nm. (Reprinted from reference with permission of the publisher.)
FIG. 23
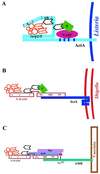
(A) Proposed model of vaccinia virus fusion with the membrane of a cell surface protrusion. (Left) Actin tail assembly pushes the viral particle against the cell membrane, forming a protrusion from the cell surface; (middle), the membrane of the viral particle fuses with the cell membrane at the tip of the protrusion; (right) the viral particle is released from the tip of the protrusion. (B) Diagram of the topology and interaction of vaccinia virus actin assembly protein A36R with other vaccinia virus membrane proteins. The cytoplasmic and luminal faces of the vaccinia virus outer IEV membrane and the amino (N) and carboxy (C) termini of A33R, A36R, A34R, and B5R are shown. (C) Diagram of SH2 and SH3 domains of the adapter protein Nck. A partial list of proteins with which these domains have been shown to interact is indicated. (Panel A adapted from reference with permission of the publisher. Panel B reprinted from reference with permission of the American Society for Microbiology. Panel C adapted from reference with permission of the publisher.)
FIG. 24
Model of actin tail assembly by vaccinia virus A36R. Nck, WIP, and N-WASP bind vaccinia A36R. Amino acid Tyr112 of vaccinia virus A36R is phosphorylated (Ph) by cellular Src family kinases. Phosphorylated A36R interacts with the adapter protein Nck. Nck, WIP, and N-WASP may exist as a preformed complex. Stimulated N-WASP activates the Arp2/3 complex. WBD, WASP-binding domain; NBD, Nck-binding domain; Pro, proline-rich region; N-WASP abbreviations as in the legend to Fig. 20.
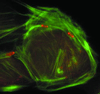
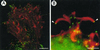
FIG. 25
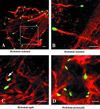
Actin tails formed by Rickettsia spp. Fluorescence microscopy of _Rickettsia_-infected Vero cells, labeled for actin (red) and Rickettsia (green). (A) Actin tails formed by spotted fever group species R. rickettsii. (B) Higher magnification of the inset from panel A, showing distinct actin bundles of the tail wrapped in a helical fashion. (C) Short hook-shaped actin tails formed by typhus group species R. typhi. (D) Absence of actin tails on typhus group species R. prowazekii. (Adapted from reference with permission of the American Society for Microbiology.)
FIG. 26
Transmission electron microscopy showing actin tails formed by R. rickettsii. (A) Long parallel filaments extending from the sides of the bacterium and absent from the bacterial pole. (B) Decoration of the filaments within the tail with the S1 subfragment of myosin, demonstrating that the barbed ends of the filaments face the bacterial body. (C) Higher magnification of filaments decorated with the S1 subfragment of myosin. The inset shows the decorated filament marked by an asterisk. Bars, 0.5 μm. (Reprinted from reference with permission of the American Society for Microbiology.)
FIG. 27
Comparison of the models of actin assembly by Listeria, Shigella, and vaccinia virus. Diagrams of molecules that interact directly or indirectly with the microbial proteins that mediate actin assembly are shown. (A) Listeria ActA; (B) Shigella IcsA; (C) vaccinia virus A36R. A, acidic domain; AB, actin-binding domain; P, profilin; Ph, phosphorylation. N-WASP abbreviations are as in the legend to Fig. 20.
Similar articles
- Molecular mechanisms of cell-cell spread of intracellular bacterial pathogens.
Ireton K. Ireton K. Open Biol. 2013 Jul 17;3(7):130079. doi: 10.1098/rsob.130079. Open Biol. 2013. PMID: 23864553 Free PMC article. Review. - Tyrosine phosphorylation is required for actin-based motility of vaccinia but not Listeria or Shigella.
Frischknecht F, Cudmore S, Moreau V, Reckmann I, Röttger S, Way M. Frischknecht F, et al. Curr Biol. 1999 Jan 28;9(2):89-92. doi: 10.1016/s0960-9822(99)80020-7. Curr Biol. 1999. PMID: 10021367 - Actin cytoskeleton. Missing link for intracellular bacterial motility?
Pollard TD. Pollard TD. Curr Biol. 1995 Aug 1;5(8):837-40. doi: 10.1016/s0960-9822(95)00167-9. Curr Biol. 1995. PMID: 7583135 - Interaction of vaccinia virus with the actin cytoskeleton.
Way M. Way M. Folia Microbiol (Praha). 1998;43(3):305-10. doi: 10.1007/BF02818616. Folia Microbiol (Praha). 1998. PMID: 9717258 Review. - Surfing pathogens and the lessons learned for actin polymerization.
Frischknecht F, Way M. Frischknecht F, et al. Trends Cell Biol. 2001 Jan;11(1):30-38. doi: 10.1016/s0962-8924(00)01871-7. Trends Cell Biol. 2001. PMID: 11146296 Review.
Cited by
- The Architecture of Iron Microbial Mats Reflects the Adaptation of Chemolithotrophic Iron Oxidation in Freshwater and Marine Environments.
Chan CS, McAllister SM, Leavitt AH, Glazer BT, Krepski ST, Emerson D. Chan CS, et al. Front Microbiol. 2016 Jun 1;7:796. doi: 10.3389/fmicb.2016.00796. eCollection 2016. Front Microbiol. 2016. PMID: 27313567 Free PMC article. - Effects of ectopically expressed neuronal Wiskott-Aldrich syndrome protein domains on Rickettsia rickettsii actin-based motility.
Harlander RS, Way M, Ren Q, Howe D, Grieshaber SS, Heinzen RA. Harlander RS, et al. Infect Immun. 2003 Mar;71(3):1551-6. doi: 10.1128/IAI.71.3.1551-1556.2003. Infect Immun. 2003. PMID: 12595475 Free PMC article. - Identification of interactions among host and bacterial proteins and evaluation of their role early during Shigella flexneri infection.
Miller KA, Garza-Mayers AC, Leung Y, Goldberg MB. Miller KA, et al. Microbiology (Reading). 2018 Apr;164(4):540-550. doi: 10.1099/mic.0.000637. Epub 2018 Feb 28. Microbiology (Reading). 2018. PMID: 29488864 Free PMC article. - Listeria monocytogenes and Shigella flexneri Activate the NLRP1B Inflammasome.
Neiman-Zenevich J, Stuart S, Abdel-Nour M, Girardin SE, Mogridge J. Neiman-Zenevich J, et al. Infect Immun. 2017 Oct 18;85(11):e00338-17. doi: 10.1128/IAI.00338-17. Print 2017 Nov. Infect Immun. 2017. PMID: 28808162 Free PMC article. - H-NS, Its Family Members and Their Regulation of Virulence Genes in Shigella Species.
Picker MA, Wing HJ. Picker MA, et al. Genes (Basel). 2016 Dec 1;7(12):112. doi: 10.3390/genes7120112. Genes (Basel). 2016. PMID: 27916940 Free PMC article. Review.
References
- Ahern-Djamali S M, Comer A R, Bachmann C, Kastenmeier A S, Reddy S K, Beckerle M C, Walter U, Hoffmann F M. Mutations in Drosophila Enabled and rescue by human vasodilator-stimulated phosphoprotein (VASP) indicate important functional roles for Ena/VASP homology domain 1 (EVH1) and EVH2 domains. Mol Biol Cell. 1998;9:2157–2171. - PMC - PubMed
- Amann K J, Pollard T D. The Arp2/3 complex nucleates actin filament branches from the sides of pre-existing filaments. Nat Cell Biol. 2001;3:306–310. - PubMed
- Anton I M, Lu W, Mayer R J, Ramesh N, Geha R S. The Wiskott-Aldrich syndrome protein-interacting protein (WIP) binds to the adaptor protein Nck. J Biol Chem. 1998;273:20992–20995. - PubMed
- Bachmann C, Fischer L, Walter U, Reinhard M. The EVH2 domain of the vasodilator-stimulated phosphoprotein mediates tetramerization, F-actin binding and actin bundle formation. J Biol Chem. 1999;274:23549–23557. - PubMed
- Bailly M, Ichetovkin I, Grant W, Zebda N, Machesky L M, Segall J E, Condeelis J. The F-actin side binding activity of the Arp2/3 complex is essential for actin nucleation and lamellipod extension. Curr Biol. 2001;11:620–625. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous