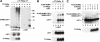PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies - PubMed (original) (raw)
PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies
S Sachdev et al. Genes Dev. 2001.
Abstract
The Wnt-responsive transcription factor LEF1 can activate transcription in association with beta-catenin and repress transcription in association with Groucho. In search of additional regulatory mechanisms of LEF1 function, we identified the protein inhibitor of activated STAT, PIASy, as a novel interaction partner of LEF1. Coexpression of PIASy with LEF1 results in potent repression of LEF1 activity and in covalent modification of LEF1 with SUMO. PIASy markedly stimulates the sumoylation of LEF1 and multiple other proteins in vivo and functions as a SUMO E3 ligase for LEF1 in a reconstituted system in vitro. Moreover, PIASy binds to nuclear matrix-associated DNA sequences and targets LEF1 to nuclear bodies, suggesting that PIASy-mediated subnuclear sequestration accounts for the repression of LEF1 activity.
Figures
Figure 1
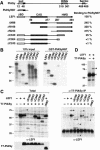
PIASy associates with LEF1 in vitro and in vivo. (A) Schematic line diagrams of PIASy and LEF1. PIASy contains a putative chromatin-binding SAP domain, a C2HC3 RING domain (RING), and a C-terminal serine-rich and acidic domain (Ser/Ac). Amino acids 1 to 97 of PIASy (PIASyN97) were sufficient for interaction with LEF1 in a yeast two-hybrid screen. LEF1 contains a β-catenin interaction domain (βBD), a context-dependent activation domain (CAD), and a high-mobility group DNA-binding domain (HMG). Numbers below the line diagrams indicate the amino acid positions of the respective protein domains. The nuclear localization signal in PIASy and in LEF1 is indicated by a black box. The ability of N- or C-terminal–truncated LEF1 proteins to associate with immobilized GST-PIASyN97 was quantified by Phosphorimager analysis and is presented as a percentage relative to the binding of full-length LEF1. (B) In vitro association of LEF1 and PIASy. N- or C-terminal–truncated LEF1 proteins were in vitro translated in the presence of [35S]-methionine, resolved by SDS-PAGE, and detected by fluorography. (Left) Ten percent of the input LEF1 proteins. Numbers indicate the molecular mass of protein markers in kilodaltons. (Right) LEF1 proteins bound to immobilized GST-PIASyN97. The in vitro translated LEF1 proteins did not bind to immobilized GST (data not shown). (C) In vivo association of LEF1 and PIASy. N- or C-terminal–truncated LEF1 and T7 epitope-tagged PIASy proteins were transiently expressed in 293T cells. Equivalent amounts of total cellular protein were immunoprecipitated with anti-T7 mAb. Coimmunoprecipitated LEF1 proteins were detected by an anti-LEF1 immunoblot (right). The arrowheads indicate LEF1 proteins; the asterisks, 18-kD-larger, modified forms of LEF1. The prominent protein at 30 kD in all lanes is IgG (right). The expression of LEF1 or T7-PIASy proteins in total cell lysates was determined by anti-LEF1 or anti-T7 immunoblots, respectively (left). PIASyΔN93 contains a deletion of amino acids 1 to 93 in PIASy and migrates faster than full-length PIASy (bottom left). (D) Covalent modification of LEF1. LEF1 proteins in total cell lysates were detected by an anti-LEF1 immunoblot. The arrowhead indicates LEF1; the asterisk, an 18-kD-larger, modified form of LEF1.
Figure 2
LEF1 is SUMO-modified in vivo. (A) PIASy associates with SUMO-modified LEF1. LEF1 and T7-PIASy were transiently expressed in 293T cells. Equivalent amounts of total cellular protein were immunoprecipitated with anti-T7 mAb. Coimmunoprecipitated endogenous, SUMO-conjugated proteins were detected by an anti-SUMO2/3 immunoblot (upper left). The anti-SUMO2/3 immunoblot was stripped and reprobed with an anti-LEF1 antibody (upper right). Numbers to the right indicate the molecular mass of protein markers in kilodaltons. The expression of LEF1 or T7-PIASy proteins in total cell lysates was determined by anti-LEF1 (middle) or anti-T7 (bottom) immunoblots, respectively. (B) LEF1 is SUMO-modified in vivo. LEF1, T7-PIASy, and Flag epitope-tagged SUMO1 or SUMO2 were transiently expressed in 293T cells. Equivalent amounts of total cellular protein were immunoprecipitated with anti-T7 mAb. Coimmunoprecipitated LEF1 proteins were detected by an anti-LEF1 immunoblot (top panel). The expression of LEF1 or T7-PIASy proteins in total cell lysates was determined by anti-LEF1 (middle) or anti-T7 (bottom) immunoblots, respectively. (C) LEF1, Flag epitope-tagged SUMO1 or SUMO2 and T7-PIASy were transiently expressed in 293T cells. LEF1 proteins in total RIPA buffer cell lysates were detected by an anti-LEF1 immunoblot (top). The expression of T7-PIASy proteins in total cell lysates was determined by an anti-T7 immunoblot (bottom).
Figure 3
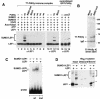
PIASy enhances SUMO-conjugation in vivo. (A) Wild-type PIASy enhances, whereas mutant PIASy proteins abrogate, SUMO-conjugation. Wild-type, RING mutant, or Ser mutant T7-PIASy proteins were coexpressed with Flag-SUMO1 or Flag-SUMO2 in 293T cells. The RING mutant contains serine substitutions for cysteines 330, 335, and 340 and an alanine substitution for histidine 337 in the RING domain. The Ser mutant contains alanine substitutions for serines 470 to 474 in the Ser/Ac domain. Flag-SUMO–modified proteins from total sample buffer cell lysates were detected by an anti-Flag immunoblot (top). Numbers to the right indicate the molecular mass of protein markers in kilodaltons. The expression of T7-PIASy proteins in total cell lysates was determined by an anti-T7 immunoblot (bottom). (B) Characterization of mutant PIASy and LEF1 proteins. Wild-type or mutant LEF1 and T7-PIASy proteins were transiently expressed in 293T cells. Equivalent amounts of total cellular protein were immunoprecipitated with anti-T7 mAb. Coimmunoprecipitated LEF1 proteins were detected by an anti-LEF1 immunoblot (top). The LEF1 M2/K267R protein contains alanine substitutions for lysine 25, aspartate 26, and glutamate 27 (Hsu et al. 1998) and an arginine substitution for lysine 267 within the two consensus SUMO motifs. The expression of LEF1 or T7-PIASy proteins in total cell lysates was determined by anti-LEF1 (middle) or anti-T7 (bottom) immunoblots, respectively.
Figure 4
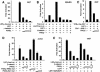
PIASy functions as a SUMO E3 ligase for LEF1 in vitro. (A) PIASy stimulates SUMO conjugation of LEF1. Recombinant LEF1 (25 ng) was incubated in the absence or presence of 5 mM ATP, 60 ng recombinant SUMO1 or SUMO2, 150 ng of recombinant E1 (Aos1/Uba2 heterodimer), 10 ng of recombinant E2 (Ubc9), and 4 μL of immune complex (IC)–purified wild-type (wt), RING mutant, or mock-transfected T7-PIASy for 60 min at 30°C, as indicated. (Lanes 14–16) Bacterially expressed wild-type GST-PIASy (300 or 1000 ng) was used instead of IC-purified T7-PIASy, as indicated. Reactions were terminated by the addition of sample buffer. LEF1 and SUMO-modified LEF1 were resolved by SDS-PAGE and detected by anti-LEF1 immunoblots. SUMO-modified LEF1 in lanes 15 and 16 migrates slower than in lanes 3 and 12 because of different electrophoresis conditions. (B) IC-purified T7-PIASy proteins were analyzed by SDS-PAGE and detected by silver staining to determine their purity and relative abundance. (C) SUMO-modified LEF1 binds DNA. Recombinant LEF1 either was sumoylated in the presence of ATP, E1, E2, SUMO1, and wild-type PIASy as described above (SUMO1-LEF1) or was treated under the same conditions without the E1 enzyme (ΔE1), so that LEF1 did not become sumoylated. The extent of LEF1 sumoylation was determined by an anti-LEF1 immunoblot using equivalent amounts of the ΔE1 and sumoylation reactions (D, input). The ability of unmodified LEF1 (25 ng) or sumoylated LEF1 (25 ng) to bind to a wt or mutated (mut) LEF1 site was determined by electrophoretic mobility shift assays. (D) SUMO-modified LEF1 binds to β-catenin. The ability of equivalent amounts of unmodified or SUMO-modified LEF1 to bind to immobilized His6–β-catenin was analyzed by an in vitro association assay. The input, unbound, and His6–β-catenin–bound LEF1 was detected by an anti-LEF1 immunoblot. The recombinant LEF1 proteins did not bind to nickel beads alone (data not shown).
Figure 5
PIASy represses LEF1 activity. (A) PIASy represses LEF1 activity from a multimerized LEF1 reporter. 293T cells were transfected with 1 μg of a LEF1 luciferase reporter construct containing multimerized LEF1-binding sites together with expression constructs encoding for β-galactosidase (25 ng; for normalization), LEF1 (30 ng), β-catenin (1 μg), or increasing amounts of T7-PIASy (0.1, 0.3, or 1 μg), as indicated. For this and subsequent experiments, the levels of luciferase or CAT activity were normalized for β-galactosidase activity and are expressed as fold activation relative to the level of luciferase or CAT activity from cells transfected with the reporter construct alone. All of the transfection experiments were performed at least three times, and the results of representative experiments are shown. (B) PIASy represses LEF1-dependent Twn reporter activity. NMuMG epithelial cells were transfected with 2 μg of the Twn luciferase reporter construct containing either wild-type (Twn-Luc) or mutated (ΔLEF-Twn-Luc) LEF1-binding sites together with expression constructs encoding for β-galactosidase (0.5 μg; for normalization), LEF1 (0.5 μg), β-catenin (5 μg), or PIASy (1 μg), as indicated. (C) PIASy represses TCRα enhancer activity. 293T cells were transfected with 0.25 μg of a TCRα-CAT reporter construct together with expression constructs encoding for β-galactosidase (50 ng; for normalization), LEF1 (100 ng), Ets1, and AML1 (250 ng each) or T7-PIASy (0.3 or 1 μg), as indicated. (D) Wild-type PIASy represses endogenous LEF1 activity. Jurkat cells were transfected with 1 μg of a LEF1 luciferase reporter construct containing multimerized LEF1-binding sites together with expression constructs encoding for β-galactosidase (0.5 μg; for normalization), β-catenin (5 μg), or increasing amounts of wild-type T7-PIASy (1 or 3 μg), RING-mutated T7-PIASy (1 or 3 μg), or Ser-mutated T7-PIASy (1 or 3 μg), as indicated. (E) Mutation of the SUMO consensus sites in LEF1 does not abrogate PIASy-mediated repression of LEF1 activity. 293T cells were transfected with 1 μg of a LEF1 luciferase reporter construct containing multimerized LEF1-binding sites together with expression constructs encoding for β-galactosidase (25 ng; for normalization), wild-type or mutant M2/K267R LEF1 (30 ng), β-catenin (1 μg), or increasing amounts of T7-PIASy (0.1, 0.3, or 1 μg), as indicated.
Figure 6
PIASy binds to nuclear matrix attachment region (MAR) DNA. (A) The PIASy SAP domain binds to MAR DNA. A GST-PIASyN97 recombinant fusion protein (0.1 or 0.3 μg) encompassing the putative SAP domain was examined for its ability to bind to a wild-type (wt) or mutated (mut) MAR consensus sequence by electrophoretic mobility shift assays, as indicated. (B) The N-terminal SAP domain of PIASy is required for nuclear matrix association in vivo. COS7 cells were transiently transfected with expression vectors encoding for wt T7-PIASy, a mutant T7-PIASy protein lacking its N-terminal 93 amino acids and SAP domain but containing a heterologous NLS for nuclear targeting (T7-PIASy–Δ93-NLS), or protein A–tagged Bright. At 48 h posttransfection, the cells were either immediately fixed (upper panels) or were processed for nuclear matrix preparations before fixation (lower panels). T7 epitope-tagged PIASy proteins were detected by indirect immunofluorescence with an anti-T7 mAb. Bright, a protein known to associate with MARs and the nuclear matrix, was detected by indirect immunofluorescence with rabbit IgG. Images were collected by confocal microscopy. The cells shown are representative of >70% of transfected cells.
Figure 7
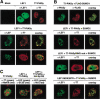
(A) PIASy relocalizes LEF1 to nuclear bodies. COS7 cells were either mock transfected or were transfected with expression constructs encoding for LEF1 or T7-PIASy. The intracellular distribution of the indicated proteins was detected by indirect immunofluorescence and analyzed by confocal microscopy. The intracellular distribution of LEF1 and T7-PIASy was detected with anti-LEF1 antibody and anti-T7 mAb, respectively. LEF1 alone displays a homogeneous distribution throughout the nucleus (upper, middle panel) but is detected in punctate nuclear bodies in the presence of T7-PIASy (middle, lower panels). Approximately 20% of the cells showed colocalization of LEF1 and T7-PIASy in <20 nuclear bodies (_middle_ panels),whereas >50% of the cells showed colocalization of LEF1 and T7-PIASy in multiple (>20) nuclear bodies (lower panels). (B) PIASy colocalizes with SUMO-modified proteins. COS7 cells were transfected with expression constructs encoding for wt T7-PIASy, T7-PIASy RING mut, Flag-SUMO1, Flag-SUMO2, wt LEF1, or mut LEF1 M2/K267R, as indicated. PIASy in the upper panels was detected with anti-PIASy antibody. The T7-PIASy proteins in the remaining panels were detected with anti-T7 mAb. Flag-SUMO1 was detected with anti-Flag mAb. T7-PIASy efficiently colocalized with Flag-SUMO1 or Flag-SUMO2 in nuclear bodies in >80% of cotransfected cells (top panels; data not shown). Coexpression of Flag-SUMO1 or Flag-SUMO2 with LEF1 and T7-PIASy enhanced nuclear body localization of LEF1 by approximately twofold (second panels; data not shown). The T7-PIASy RING mutant protein displays homogeneous staining throughout the nucleus and is unable to relocalize LEF1 to nuclear bodies (third panels). The LEF1 M2/K267R mutant protein is efficiently relocalized to nuclear bodies in the presence of T7-PIASy and Flag-SUMO2 (bottom panels). (C) PIASy partially colocalizes with PML bodies. T7-PIASy, LEF1, and epitope-tagged HA-Sp100 were transfected into COS7 cells, as indicated. The intracellular distributions of T7-PIASy and dimethyl-lysine 9 of histone H3, a marker for heterochromatin, were detected with anti-T7 mAb and anti-MeK9, respectively (left). The distribution of LEF1 (coexpressed with T7-PIASy) and SC35, a component of the splicing machinery, was detected with anti-LEF1 and anti-SC35 mAb, respectively (center). The distribution of LEF1 (coexpressed with T7-PIASy) and HA-Sp100, a component of PML nuclear bodies, was detected with anti-LEF1 antibody and anti-HA mAb, respectively (right). LEF1 only showed a partially overlapping pattern of distribution with HA-Sp100 in PML nuclear bodies.
Comment in
- A new RING for SUMO: wrestling transcriptional responses into nuclear bodies with PIAS family E3 SUMO ligases.
Jackson PK. Jackson PK. Genes Dev. 2001 Dec 1;15(23):3053-8. doi: 10.1101/gad.955501. Genes Dev. 2001. PMID: 11731472 Review. No abstract available.
Similar articles
- A new RING for SUMO: wrestling transcriptional responses into nuclear bodies with PIAS family E3 SUMO ligases.
Jackson PK. Jackson PK. Genes Dev. 2001 Dec 1;15(23):3053-8. doi: 10.1101/gad.955501. Genes Dev. 2001. PMID: 11731472 Review. No abstract available. - Modification of GATA-2 transcriptional activity in endothelial cells by the SUMO E3 ligase PIASy.
Chun TH, Itoh H, Subramanian L, Iñiguez-Lluhí JA, Nakao K. Chun TH, et al. Circ Res. 2003 Jun 13;92(11):1201-8. doi: 10.1161/01.RES.0000076893.70898.36. Epub 2003 May 15. Circ Res. 2003. PMID: 12750312 - SUMO-1 modification of PIASy, an E3 ligase, is necessary for PIASy-dependent activation of Tcf-4.
Ihara M, Yamamoto H, Kikuchi A. Ihara M, et al. Mol Cell Biol. 2005 May;25(9):3506-18. doi: 10.1128/MCB.25.9.3506-3518.2005. Mol Cell Biol. 2005. PMID: 15831457 Free PMC article. - Identification of a new small ubiquitin-like modifier (SUMO)-interacting motif in the E3 ligase PIASy.
Kaur K, Park H, Pandey N, Azuma Y, De Guzman RN. Kaur K, et al. J Biol Chem. 2017 Jun 16;292(24):10230-10238. doi: 10.1074/jbc.M117.789982. Epub 2017 Apr 28. J Biol Chem. 2017. PMID: 28455449 Free PMC article. - Novel initiation genes in squamous cell carcinomagenesis: a role for substrate-specific ubiquitylation in the control of cell survival.
Albor A, Kulesz-Martin M. Albor A, et al. Mol Carcinog. 2007 Aug;46(8):585-90. doi: 10.1002/mc.20344. Mol Carcinog. 2007. PMID: 17626251 Review.
Cited by
- The deubiquitinase USP36 promotes snoRNP group SUMOylation and is essential for ribosome biogenesis.
Ryu H, Sun XX, Chen Y, Li Y, Wang X, Dai RS, Zhu HM, Klimek J, David L, Fedorov LM, Azuma Y, Sears RC, Dai MS. Ryu H, et al. EMBO Rep. 2021 Jun 4;22(6):e50684. doi: 10.15252/embr.202050684. Epub 2021 Apr 14. EMBO Rep. 2021. PMID: 33852194 Free PMC article. - The many faces and functions of β-catenin.
Valenta T, Hausmann G, Basler K. Valenta T, et al. EMBO J. 2012 Jun 13;31(12):2714-36. doi: 10.1038/emboj.2012.150. Epub 2012 May 22. EMBO J. 2012. PMID: 22617422 Free PMC article. Review. - Regulation of the nucleocytoplasmic trafficking of viral and cellular proteins by ubiquitin and small ubiquitin-related modifiers.
Wang YE, Pernet O, Lee B. Wang YE, et al. Biol Cell. 2012 Mar;104(3):121-38. doi: 10.1111/boc.201100105. Epub 2011 Dec 28. Biol Cell. 2012. PMID: 22188262 Free PMC article. Review. - Association with class IIa histone deacetylases upregulates the sumoylation of MEF2 transcription factors.
Grégoire S, Yang XJ. Grégoire S, et al. Mol Cell Biol. 2005 Mar;25(6):2273-87. doi: 10.1128/MCB.25.6.2273-2287.2005. Mol Cell Biol. 2005. PMID: 15743823 Free PMC article. - MafG sumoylation is required for active transcriptional repression.
Motohashi H, Katsuoka F, Miyoshi C, Uchimura Y, Saitoh H, Francastel C, Engel JD, Yamamoto M. Motohashi H, et al. Mol Cell Biol. 2006 Jun;26(12):4652-63. doi: 10.1128/MCB.02193-05. Mol Cell Biol. 2006. PMID: 16738329 Free PMC article.
References
- Aravind L, Koonin EV. aSAP: A putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci. 2000;25:112–114. - PubMed
- Azuma Y, Tan SH, Cavenagh MM, Ainsztein AM, Saitoh H, Dasso M. Expression and regulation of the mammalian SUMO-1 E1 enzyme. FASEB J. 2001;15:1825–1827. - PubMed
- Bienz M, Clevers H. Linking colorectal cancer to Wnt signaling. Cell. 2000;103:311–320. - PubMed
- Bode J, Kohwi Y, Dickinson L, Joh T, Klehr D, Mielke C, Kohwi-Shigematsu T. Biological significance of unwinding capability of nuclear matrix–associating DNAs. Science. 1992;255:195–197. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases