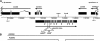c(3)G encodes a Drosophila synaptonemal complex protein - PubMed (original) (raw)
c(3)G encodes a Drosophila synaptonemal complex protein
S L Page et al. Genes Dev. 2001.
Abstract
The meiotic mutant c(3)G (crossover suppressor on 3 of Gowen) abolishes both synaptonemal complex (SC) formation and meiotic recombination, whereas mutations in the mei-W68 and mei-P22 genes prevent recombination but allow normal SC to form. These data, as well as a century of cytogenetic studies, support the argument that meiotic recombination between homologous chromosomes in Drosophila females requires synapsis and SC formation. We have cloned the c(3)G gene and shown that it encodes a protein that is structurally similar to SC proteins from yeast and mammals. Immunolocalization of the C(3)G protein, as well as the analysis of a C(3)G-eGFP expression construct, reveals that C(3)G is present in a thread-like pattern along the lengths of chromosomes in meiotic prophase, consistent with a role as an SC protein present on meiotic bivalents. The availability of a marker for SC in Drosophila allowed the investigation of the extent of synapsis in exchange-defective mutants. These studies indicate that SC formation is impaired in certain meiotic mutants and that the synaptic defect correlates with the exchange defects. Moreover, the observation of interference among the residual exchanges in these mutant oocytes implies that complete SC formation is not required for crossover interference in Drosophila.
Figures
Figure 1
The c(3)G (CG17604) locus. (A) The genomic structure of c(3)G (CG17604), based on the GM04379 and LD07655 cDNAs (Rubin et al. 2000); surrounding genes are shown as boxes representing exons. Genomic DNA coordinates in base pairs are from the Gadfly Drosophila genome annotation database (FlyBase 1999). The c(3)G (CG17604) open reading frame is shown in gray. c(3)G mutations are represented below the intron/exon structure of CG17604. CG18505 is encoded on the opposite strand as c(3)G, largely within the c(3)G 5′ untranslated region. (B) Rescue constructs P{X203} and P{X204} include genomic DNA from between the _Xba_I and _Xho_I sites indicated in A. P{X204} bears a deletion between the _Hin_dIII sites (shown in A) within the CG17604 gene.
Figure 2
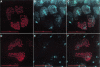
C(3)G protein colocalizes in a thread-like pattern associated with nuclear DNA. (A) Anti-C(3)G immunofluorescence (red) in an early germ-line cyst (germarium region 2a), showing a thread-like pattern visible in four nuclei. The lower two adjacent nuclei are more intensely stained and are probably the two pro-oocytes. (B) Nuclear DNA in the cyst shown in A visualized by DAPI staining (cyan). (C) Merged image of A and B, showing colocalization of C(3)G (red) with the DNA (cyan). (D) C(3)G–eGFP (pseudocolored red) localizes in a thread-like pattern similar to the pattern visualized by anti-C(3)G immunofluorescence. Two brightly fluorescing nuclei are visible in the center of the frame, and two dimly fluorescing nuclei are positioned at the bottom and upper right edges of the frame. (E) Nuclear DNA in the cyst shown in D visualized by DAPI staining (cyan). (F) Merged image of D and E, showing colocalization of C(3)G–eGFP (red) with the DNA (cyan). Bars, 10 μm.
Figure 3
C(3)G protein localizes between paired homologous chromosomes. (A) Deconvolved optical section through a pro-oocyte nucleus showing C(3)G localization (red). (B) DAPI image showing DNA (cyan) of the nucleus shown in A. Much of the chromatin is visible as paired linear tracks of staining (arrows). (C) Merged image of A and B, showing the association of the thread-like localization of C(3)G (red) to the paired linear tracks of DNA (cyan). In parts of this section, C(3)G can be observed between the tracks of DNA (arrows). Twisting of the chromosomes and/or position of the chromosomes within the section prevents the visualization of this arrangement of C(3)G and chromatin throughout the nucleus. A_–_C, bar, 5 μm. (D) Deconvolved optical section showing fluorescence in situ hybridization (FISH) of an _X_-chromosome probe (red) to an immunostained germarium. Pairs of FISH signals, representing paired homologous chromosomes, are visible in two cells (arrows). (E) Anti-C(3)G immunofluorescence (cyan) superimposed with the FISH signals (red) from D. The paired FISH signals are arranged on either side of the thread-like C(3)G immunofluorescence signal (arrows). D_–_E, bar, 10 μm.
Figure 4
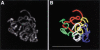
Computer model of C(3)G localization. (A) Maximum intensity projection of all anti-C(3)G immunofluorescence signals from one nucleus (lower left nucleus from Fig. 2A). (B) Wire frame model of the anti-C(3)G immunofluorescence signals shown in A. The contiguous thread-like immunofluorescence signals were modeled in three dimensions using the deconvolved image stack used to create Figure 5A. Each contiguous segment of the model is shown in a different color. Bar, 5 μm.
Figure 5
Comparison of C(3)G and Orb localization in wild-type and egl germaria. (A) Maximum intensity projection of a deconvolved image stack containing an entire w1118 germarium. Anti-C(3)G immunofluorescence (red) is visible in a subset of nuclei (visualized by DAPI staining, blue). (B) Comparison of C(3)G (red) and Orb localization (green) in the germarium shown in A. C(3)G is expressed in germ-line cells beginning at the time when Orb is first detected in the cytoplasm of region 2a cysts. In early cysts, C(3)G is detected in ∼4 cells per cyst. By region 2b, only two cells retain C(3)G, one of which is the oocyte, as determined by the accumulation of Orb protein. C(3)G is then lost from the other pro-oocyte by the time cysts leave the germarium. (C) Germarium from A, showing C(3)G (red), Orb (green), and DNA (blue). Regions 1, 2a, 2b, and 3 of the germarium are indicated at left. (D) Maximum intensity projection of a deconvolved image stack containing an entire germarium from a female homozygous for egl1. Anti-C(3)G immunofluorescence (red) is visible in numerous nuclei (visualized by DAPI staining, blue) in region 2a and early region 2b, in contrast to wild type. (E) Comparison of C(3)G (red) and Orb localization (green) in the germarium shown in D. Orb protein is expressed and is present in the cytoplasm of germ-line cells starting in region 2a, but Orb fails to accumulate in the oocyte cytoplasm, because no oocyte is determined in egl mutant ovaries. C(3)G is present in the nuclei of all 16 cells of the germ-line cyst starting in region 2a, at the same time that Orb becomes detectable. By late region 2b, all 16 cells lose anti-C(3)G staining. (F) Germarium from D, showing C(3)G (red), Orb (green), and DNA (blue).
Figure 6
Localization of C(3)G in vitellarial cysts from w1118 females. (A) Deconvolved optical section showing C(3)G (red) in a stage 3 oocyte nucleus. The Orb protein (green) is accumulated in the oocyte cytoplasm. C(3)G remains localized in thread-like patterns associated with DNA (blue) at this stage. (B) Deconvolved optical section showing C(3)G (red) in a stage 4 oocyte nucleus. Orb protein and DNA are stained as in A. Thread-like C(3)G colocalization with DNA is visible, but is beginning to dissipate. (C) Deconvolved optical section showing C(3)G (red) in a stage 6 oocyte nucleus. Orb protein and DNA are stained as in A. C(3)G localization to chromatin is decreased, and hazy extrachromosomal anti-C(3)G immunofluorescence is starting to become visible in the nucleus. In A_–_C, the posterior of each egg chamber is to the right. Bar, 10 μm.
Figure 7
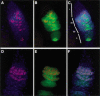
Localization of C(3)G in recombination-defective mutants. (A_–_C) Maximum intensity projections of anti-C(3)G immunofluorescence in three pro-oocyte nuclei from w1118 females. (D_–_F) Maximum intensity projections of anti-C(3)G immunofluorescence in three pro-oocyte nuclei from P{X204}/+; c(3)G68/c(3)G68 females. The mutant form of C(3)G encoded by the P{X204} construct localizes to chromosomes (data not shown), but forms an increased number of stained segments suggestive of incomplete synapsis. (G_–_I) Maximum intensity projections of anti-C(3)G immunofluorescence in three pro-oocyte nuclei from mei-P261 homozygous females. Pro-oocytes in mei-P261 germaria were identified by cytoplasmic accumulation of Orb protein (data not shown). C(3)G forms spots and short segments of linear localization, indicating an extensive lack of normal SC.
Similar articles
- [Synaptonemal complex proteins: specific proteins of meiotic chromosomes].
Penkina MV, Karpova OI, Bogdanov IuF. Penkina MV, et al. Mol Biol (Mosk). 2002 May-Jun;36(3):397-407. Mol Biol (Mosk). 2002. PMID: 12068623 Review. Russian. - [Cytogenetic patterns in synapsis of meiotic chromosomes in animals and plants].
Bogdanov IuF, Grishaeva TM, Kolomiets OL, Fedotova IuS. Bogdanov IuF, et al. Genetika. 1996 Nov;32(11):1474-93. Genetika. 1996. PMID: 9119209 Review. Russian. - Corona is required for higher-order assembly of transverse filaments into full-length synaptonemal complex in Drosophila oocytes.
Page SL, Khetani RS, Lake CM, Nielsen RJ, Jeffress JK, Warren WD, Bickel SE, Hawley RS. Page SL, et al. PLoS Genet. 2008 Sep 19;4(9):e1000194. doi: 10.1371/journal.pgen.1000194. PLoS Genet. 2008. PMID: 18802461 Free PMC article. - When specialized sites are important for synapsis and the distribution of crossovers.
Joyce EF, McKim KS. Joyce EF, et al. Bioessays. 2007 Mar;29(3):217-26. doi: 10.1002/bies.20531. Bioessays. 2007. PMID: 17295219 Review.
Cited by
- Separable Crossover-Promoting and Crossover-Constraining Aspects of Zip1 Activity during Budding Yeast Meiosis.
Voelkel-Meiman K, Johnston C, Thappeta Y, Subramanian VV, Hochwagen A, MacQueen AJ. Voelkel-Meiman K, et al. PLoS Genet. 2015 Jun 26;11(6):e1005335. doi: 10.1371/journal.pgen.1005335. eCollection 2015 Jun. PLoS Genet. 2015. PMID: 26114667 Free PMC article. - Early development of Drosophila embryos requires Smc5/6 function during oogenesis.
Tran M, Tsarouhas V, Kegel A. Tran M, et al. Biol Open. 2016 Jul 15;5(7):928-41. doi: 10.1242/bio.019000. Biol Open. 2016. PMID: 27288507 Free PMC article. - A molecular cell biology toolkit for the study of meiosis in the silkworm Bombyx mori.
Xiang Y, Tsuchiya D, Guo F, Gardner J, McCroskey S, Price A, Tromer EC, Walters JR, Lake CM, Hawley RS. Xiang Y, et al. G3 (Bethesda). 2023 May 2;13(5):jkad058. doi: 10.1093/g3journal/jkad058. G3 (Bethesda). 2023. PMID: 36911915 Free PMC article. - The cohesion protein ORD is required for homologue bias during meiotic recombination.
Webber HA, Howard L, Bickel SE. Webber HA, et al. J Cell Biol. 2004 Mar 15;164(6):819-29. doi: 10.1083/jcb.200310077. Epub 2004 Mar 8. J Cell Biol. 2004. PMID: 15007062 Free PMC article. - mei-P22 encodes a chromosome-associated protein required for the initiation of meiotic recombination in Drosophila melanogaster.
Liu H, Jang JK, Kato N, McKim KS. Liu H, et al. Genetics. 2002 Sep;162(1):245-58. doi: 10.1093/genetics/162.1.245. Genetics. 2002. PMID: 12242237 Free PMC article.
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. - PubMed
- Agard DA, Hiraoka Y, Shaw P, Sedat JW. Fluorescence microscopy in three dimensions. Methods Cell Biol. 1989;30:353–377. - PubMed
- Agarwal S, Roeder GS. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell. 2000;102:245–255. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases