Trimming, weighting, and grouping SNPs in human case-control association studies - PubMed (original) (raw)
Trimming, weighting, and grouping SNPs in human case-control association studies
J Hoh et al. Genome Res. 2001 Dec.
Abstract
The search for genes underlying complex traits has been difficult and often disappointing. The main reason for these difficulties is that several genes, each with rather small effect, might be interacting to produce the trait. Therefore, we must search the whole genome for a good chance to find these genes. Doing this with tens of thousands of SNP markers, however, greatly increases the overall probability of false-positive results, and current methods limiting such error probabilities to acceptable levels tend to reduce the power of detecting weak genes. Investigating large numbers of SNPs inevitably introduces errors (e.g., in genotyping), which will distort analysis results. Here we propose a simple strategy that circumvents many of these problems. We develop a set-association method to blend relevant sources of information such as allelic association and Hardy-Weinberg disequilibrium. Information is combined over multiple markers and genes in the genome, quality control is improved by trimming, and an appropriate testing strategy limits the overall false-positive rate. In contrast to other available methods, our method to detect association to sets of SNP markers in different genes in a real data application has shown remarkable success.
Figures
Figure 1
Flow diagram illustrating the algorithm implemented in the set-association approach.
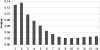
Figure 2
Significance level of Sn statistic as a function of the number n of SNPs in different genes that are included at each step. The smallest significance level, min_n_ pn, occurs with 10 SNPs included in Sn. The 10 SNPs represent 9 different genes.
Similar articles
- Quantification of the power of Hardy-Weinberg equilibrium testing to detect genotyping error.
Cox DG, Kraft P. Cox DG, et al. Hum Hered. 2006;61(1):10-4. doi: 10.1159/000091787. Epub 2006 Mar 1. Hum Hered. 2006. PMID: 16514241 - Detecting susceptibility genes in case-control studies using set association.
Kim S, Zhang K, Sun F. Kim S, et al. BMC Genet. 2003 Dec 31;4 Suppl 1(Suppl 1):S9. doi: 10.1186/1471-2156-4-S1-S9. BMC Genet. 2003. PMID: 14975077 Free PMC article. - Detection of genotyping errors and pseudo-SNPs via deviations from Hardy-Weinberg equilibrium.
Leal SM. Leal SM. Genet Epidemiol. 2005 Nov;29(3):204-14. doi: 10.1002/gepi.20086. Genet Epidemiol. 2005. PMID: 16080207 Free PMC article. - Tag SNP selection for association studies.
Stram DO. Stram DO. Genet Epidemiol. 2004 Dec;27(4):365-74. doi: 10.1002/gepi.20028. Genet Epidemiol. 2004. PMID: 15372618 Review. - SNP judgments and freedom of association.
Hegele RA. Hegele RA. Arterioscler Thromb Vasc Biol. 2002 Jul 1;22(7):1058-61. doi: 10.1161/01.atv.0000026801.56080.14. Arterioscler Thromb Vasc Biol. 2002. PMID: 12117714 Review. No abstract available.
Cited by
- Genetics of the connectome.
Thompson PM, Ge T, Glahn DC, Jahanshad N, Nichols TE. Thompson PM, et al. Neuroimage. 2013 Oct 15;80:475-88. doi: 10.1016/j.neuroimage.2013.05.013. Epub 2013 May 21. Neuroimage. 2013. PMID: 23707675 Free PMC article. Review. - The pharmacogenetics of lithium response depends upon clinical co-morbidity.
Bremer T, Diamond C, McKinney R, Shehktman T, Barrett TB, Herold C, Kelsoe JR. Bremer T, et al. Mol Diagn Ther. 2007;11(3):161-70. doi: 10.1007/BF03256238. Mol Diagn Ther. 2007. PMID: 17570738 - The p53MH algorithm and its application in detecting p53-responsive genes.
Hoh J, Jin S, Parrado T, Edington J, Levine AJ, Ott J. Hoh J, et al. Proc Natl Acad Sci U S A. 2002 Jun 25;99(13):8467-72. doi: 10.1073/pnas.132268899. Epub 2002 Jun 19. Proc Natl Acad Sci U S A. 2002. PMID: 12077306 Free PMC article. - Genome-wide conditional search for epistatic disease-predisposing variants in human association studies.
Wang G, Yang Y, Ott J. Wang G, et al. Hum Hered. 2010;70(1):34-41. doi: 10.1159/000293722. Epub 2010 Apr 23. Hum Hered. 2010. PMID: 20413980 Free PMC article. - Association study of GSK3 gene polymorphisms with schizophrenia and clozapine response.
Souza RP, Romano-Silva MA, Lieberman JA, Meltzer HY, Wong AH, Kennedy JL. Souza RP, et al. Psychopharmacology (Berl). 2008 Oct;200(2):177-86. doi: 10.1007/s00213-008-1193-9. Epub 2008 May 26. Psychopharmacology (Berl). 2008. PMID: 18500637
References
- Blangero J, Williams JT, Almasy L. Variance components methods for detecting complex trait loci. In: Rao DC, editor. Advances in genetics. Vol. 42. San Diego: Academic Press; 2000. pp. 151–181. - PubMed
- Collins FS, Brooks LD, Chakravarti A. A DNA polymorphism discovery resource for research on human genetic variation. Genome Res. 1998;8:1229–1231. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources