GeneLynx: a gene-centric portal to the human genome - PubMed (original) (raw)
GeneLynx: a gene-centric portal to the human genome
B Lenhard et al. Genome Res. 2001 Dec.
Abstract
GeneLynx is a meta-database providing an extensive collection of hyperlinks to human gene-specific information in diverse databases available on the Internet. The GeneLynx project is based on the simple notion that given any gene-specific identifier (accession number, gene name, text, or sequence), scientists should be able to access a single location that provides a set of links to all the publicly available information pertinent to the specified human gene. GeneLynx was implemented as an extensible relational database with an intuitive and user-friendly Web interface. The data are automatically extracted from more than 40 external resources, using appropriate approaches to maximize coverage of the available data. Construction and curation of the system is mediated by a custom set of software tools. An indexing utility is provided to facilitate the establishment of hyperlinks in external databases. A unique feature of the GeneLynx system is a communal curation system for user-aided annotation. GeneLynx can be accessed freely at http://www.genelynx.org.
Figures
Figure 1
A screenshot of the GeneLynx record Web page, containing a comprehensive set of links for the given gene (hypoxanthine phosphoribosyltransferase 1 is shown).
Figure 2
A screenshot of the GeneLynx user comment submission interface. The submitted comment, together with the curator's response, is available for users to consider.
Figure 3
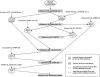
A scheme of a subset of the associations-building procedure used for the construction of GeneLynx database. The central (shaded) items are the associations between GeneLynx and other resources.
Figure 4
A screenshot of the Resolver curation tool for resolving ambiguous associations. The one-to-many relationship between a GeneLynx cDNA cluster and associated UniGene clusters is resolved by inspecting an array of dot plots. The curator can, if necessary, consult NCBI records for each cDNA and UniGene cluster, which are accessed via provided hyperlinks.
Similar articles
- GeneLynx mouse: integrated portal to the mouse genome.
Lenhard B, Wahlestedt C, Wasserman WW. Lenhard B, et al. Genome Res. 2003 Jun;13(6B):1501-4. doi: 10.1101/gr.951403. Genome Res. 2003. PMID: 12819149 Free PMC article. - The Gene Set Builder: collation, curation, and distribution of sets of genes.
Yusuf D, Lim JS, Wasserman WW. Yusuf D, et al. BMC Bioinformatics. 2005 Dec 21;6:305. doi: 10.1186/1471-2105-6-305. BMC Bioinformatics. 2005. PMID: 16371163 Free PMC article. - GeneCruiser: a web service for the annotation of microarray data.
Liefeld T, Reich M, Gould J, Zhang P, Tamayo P, Mesirov JP. Liefeld T, et al. Bioinformatics. 2005 Sep 15;21(18):3681-2. doi: 10.1093/bioinformatics/bti587. Epub 2005 Jul 19. Bioinformatics. 2005. PMID: 16030072 - The Pain Genes Database: An interactive web browser of pain-related transgenic knockout studies.
Lacroix-Fralish ML, Ledoux JB, Mogil JS. Lacroix-Fralish ML, et al. Pain. 2007 Sep;131(1-2):3.e1-4. doi: 10.1016/j.pain.2007.04.041. Epub 2007 Jun 14. Pain. 2007. PMID: 17574758 Review. - LinkHub: a Semantic Web system that facilitates cross-database queries and information retrieval in proteomics.
Smith AK, Cheung KH, Yip KY, Schultz M, Gerstein MK. Smith AK, et al. BMC Bioinformatics. 2007 May 9;8 Suppl 3(Suppl 3):S5. doi: 10.1186/1471-2105-8-S3-S5. BMC Bioinformatics. 2007. PMID: 17493288 Free PMC article. Review.
Cited by
- AbsIDconvert: an absolute approach for converting genetic identifiers at different granularities.
Mohammad F, Flight RM, Harrison BJ, Petruska JC, Rouchka EC. Mohammad F, et al. BMC Bioinformatics. 2012 Sep 12;13:229. doi: 10.1186/1471-2105-13-229. BMC Bioinformatics. 2012. PMID: 22967011 Free PMC article. - The UCSC Genome Browser.
Karolchik D, Hinrichs AS, Kent WJ. Karolchik D, et al. Curr Protoc Bioinformatics. 2009 Dec;Chapter 1:Unit1.4. doi: 10.1002/0471250953.bi0104s28. Curr Protoc Bioinformatics. 2009. PMID: 19957273 Free PMC article. - Exploring the foundation of genomics: a northern blot reference set for the comparative analysis of transcript profiling technologies.
Kemmer D, Faxén M, Hodges E, Lim J, Herzog E, Ljungström E, Lundmark A, Olsen MK, Podowski R, Sonnhammer EL, Nilsson P, Reimers M, Lenhard B, Roberds SL, Wahlestedt C, Höög C, Agarwal P, Wasserman WW. Kemmer D, et al. Comp Funct Genomics. 2004;5(8):584-95. doi: 10.1002/cfg.443. Comp Funct Genomics. 2004. PMID: 18629180 Free PMC article. - MADIBA: a web server toolkit for biological interpretation of Plasmodium and plant gene clusters.
Law PJ, Claudel-Renard C, Joubert F, Louw AI, Berger DK. Law PJ, et al. BMC Genomics. 2008 Feb 28;9:105. doi: 10.1186/1471-2164-9-105. BMC Genomics. 2008. PMID: 18307768 Free PMC article. - Gene characterization index: assessing the depth of gene annotation.
Kemmer D, Podowski RM, Yusuf D, Brumm J, Cheung W, Wahlestedt C, Lenhard B, Wasserman WW. Kemmer D, et al. PLoS One. 2008 Jan 23;3(1):e1440. doi: 10.1371/journal.pone.0001440. PLoS One. 2008. PMID: 18213364 Free PMC article.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
- Etzold T, Ulyanov A, Argos P. SRS: Information retrieval system for molecular biology data banks. Methods Enzymol. 1996;266:114–128. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical