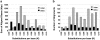Human-specific duplication and mosaic transcripts: the recent paralogous structure of chromosome 22 - PubMed (original) (raw)
Comparative Study
doi: 10.1086/338458. Epub 2001 Nov 30.
Affiliations
- PMID: 11731936
- PMCID: PMC419985
- DOI: 10.1086/338458
Comparative Study
Human-specific duplication and mosaic transcripts: the recent paralogous structure of chromosome 22
Jeffrey A Bailey et al. Am J Hum Genet. 2002 Jan.
Abstract
In recent decades, comparative chromosomal banding, chromosome painting, and gene-order studies have shown strong conservation of gross chromosome structure and gene order in mammals. However, findings from the human genome sequence suggest an unprecedented degree of recent (<35 million years ago) segmental duplication. This dynamism of segmental duplications has important implications in disease and evolution. Here we present a chromosome-wide view of the structure and evolution of the most highly homologous duplications (> or = 1 kb and > or = 90%) on chromosome 22. Overall, 10.8% (3.7/33.8 Mb) of chromosome 22 is duplicated, with an average sequence identity of 95.4%. To organize the duplications into tractable units, intron-exon structure and well-defined duplication boundaries were used to define 78 duplicated modules (minimally shared evolutionary segments) with 157 copies on chromosome 22. Analysis of these modules provides evidence for the creation or modification of 11 novel transcripts. Comparative FISH analyses of human, chimpanzee, gorilla, orangutan, and macaque reveal qualitative and quantitative differences in the distribution of these duplications--consistent with their recent origin. Several duplications appear to be human specific, including a approximately 400-kb duplication (99.4%-99.8% sequence identity) that transposed from chromosome 14 to the most proximal pericentromeric region of chromosome 22. Experimental and in silico data further support a pericentromeric gradient of duplications where the most recent duplications transpose adjacent to the centromere. Taken together, these data suggest that segmental duplications have been an ongoing process of primate genome evolution, contributing to recent gene innovation and the dynamic transformation of genome architecture within and among closely related species.
Figures
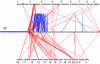
Figure 1
Spatial distribution of large segmental duplications between chromosome 22 and other human chromosomes. A scaled (50×) version of chromosome 22, surrounded by the other chromosomes, shows lines representing interchromosomal (red) and intrachromosomal (blue) alignments (⩾10 kb). The majority of chromosome 22 pericentromeric duplications localize to the pericentromeric regions of other chromosomes. Likewise, the majority of subtelomeric duplications localize to subtelomeric regions of nonhomologous chromosomes. There is little cross-hybridization between subtelomeric and pericentromeric duplications. Chromosomes 2 and 20 share the largest amount of sequence with chromosome 22, whereas chromosomes 5, 7, 14, 18, 19, and X do not share any duplications with chromosome 22 that are >10 kb in size. The coordinates are based on the published UCSC human genome assembly. For chromosome 22, each tick mark represents a 1-Mb interval. For the other chromosomes, tick marks represent 50-Mb intervals. Purple boxes represent the unsequenced centromeres, acrocentric p arms, and Y heterochromatin. Gaps are denoted by white space. The program PARASIGHT was used to generate this diagram.
Figure 2
Pairwise sequence distance (K) of chromosome 22 duplications. The two histograms show the distribution of genetic distance in terms of the number of aligned base pairs (a) and the number of alignments (b). Alignments are separated into interchromosomal (gray) and intrachromosomal (black). Distance (K) is the number of substitutions per 100 bp aligned and was corrected for multiple substitutions (see Material and Methods section).
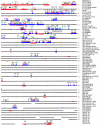
Figure 3
Interchromosomal duplications of the pericentromeric region of chromosome 22. The combined results of in silico and FISH duplication detection are displayed for the most proximal 2 Mb of 22q. Labeled dark gray boxes above the tick-marked sequence denote the positions of chromosome 22 clones used for FISH analysis. Below the sequence, light gray boxes represent positive FISH signals to particular chromosomes. Black bars show the in silico positions of duplicated alignments, on the basis of comparison of the chromosome 22 reference sequence to the rest of the human genome. The majority of the paralogous segments mapped to pericentromeric positions on the other chromosomes. CER denotes a region containing a 150-kb expanse of centromeric-associated repeat. Blank spaces represent sequence gaps. “UK” denotes sequence with unknown chromosome assignments.
Figure 4
Comparative FISH of a human-specific duplication. Human, chimpanzee, and gorilla comparative FISH results are shown for human chromosome 22 probes 140m6 (a) and 134c5 (b). Both BACs lack any signal to chromosome 22 among the nonhuman primates.
Figure 5
PCR analysis of a 550-kb duplication between human chromosomes 14 and 22. The figure shows the position on chromosome 22 of oligonucleotides that were designed to amplify paralogous sequences from chromosomes 14 and 22. Ten PCR products (A-K) were designed, spanning ∼550 kb. Products were amplified and sequenced from both chromosome 14 and chromosome 22. The total high-quality sequence (SEQ TOTAL), the number of sites with fixed differences (SEQ fixed) between 22 and 14, and the number of heterogeneous sites (SEQ hetero) for each product are shown. These heterogeneous sites in the most centromeric products suggest multiple copies for this region within chromosome 14. The average sequence identity between chromosome 14 and 22 for all 3,215 bases was 99.4%–99.8% (with and without heterogeneous sites).
Figure 6
The modular structure of segmental duplications on chromosome 22. The position and size of the 78 defined modules are shown along the entire chromosome 22 sequence (black line; each line = 1 Mb). Modules are arbitrarily colored, except that gray and black are used for interchromosomal duplications. Arrows indicate orientation relative either to a defining transcript or to the most proximal copy. The positions of interchromosomal (red bars) and intrachromosomal (blue bars) duplications are shown overlapping the sequence line. Tick marks represent 100 kb. Gaps (white space) in the sequence are drawn to scale. The program PARASIGHT was used to generate this diagram.
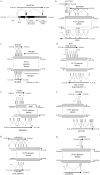
Figure 7
Transcripts created or modified through segmental duplication. We identified 11 transcripts that have been created or modified via the process of segmental duplication. This was a comprehensive and stringent search of chromosome 22 duplications, to identify overlapping regions of transcriptional activity. Transcriptional activity was based on finding two or more spliced cDNA sequences that had been placed to their best genomic location (see Material and Methods section). Eight examples illustrating the intron-exon structure, as well as the underlying duplications, are shown for the new (top) and putative ancestral (bottom) transcripts. Positions within the genome assembly are given in kb. Exons are positioned approximately, but exon size is not shown to scale. a, AL001299, a full-length transcript (1,625 bases) that originates from mosaic modules within the pericentromeric region. It has a putative ORF of 98 aa. The intron-exon structure spans ∼100 kb (14,027–14,124 kb), with each exon originating from a different module. Two modules underlying the gene show expressed genes suggesting the ancestral origin of these modules: solute carrier family 25 member 15 (SLC25A15), for the 13q14 module, and von Willebrand factor (vWF), for the 12p11 module. Thus, the pericentromeric juxtaposition of these modules leads to the formation of AL001299. Exon 2 does not contain any exon sequence from SLC25A15. Exon 3 is composed of vWF exon sequence, albeit in the reverse orientation. b, Partial-gene duplication of the proximal seven exons of lipoprotein receptor–related protein 5 (LRP5) from 11q13. Alignment of five transcripts suggests multiple transcriptional start sites or alternative splicing. Both AL137651 and AI972731 utilize exon sequence from LRP5, including exons 1, 4, 5, 6, 7, 8, and 9. The best ORFs are 252 aa for AL137651 and 77 aa for BE396696. c, Whole-gene duplication (ancestral copy undetermined) leading to the formation of DGCR6 and DGCR6L genes. The duplication also includes a whole-gene duplication of proline dehydrogenase (PRODH), which forms an unprocessed pseudogene (PRODHΨ) in the distal copy. DGCR6 and DGCR6L transcripts have conserved intron-exon and coding structure (220 aa). The transcripts have been experimentally verified and show expression from multiple tissues, with differential expression between the two copies (Edelmann et al. 2001). Function is unknown. d, Partial-gene duplication of the seven terminal exons (17–23) of BCR (NM_021574) that has led to the creation of a fusion transcript in one of the distal copies. The full-length transcript (NM_014549) has seven exons and is in the reverse orientation, compared to BCR. Exon 1 is derived from the flanking distal chromosome 22 sequence and exons 2–7 are derived from the duplicated sequence. These terminal exons incorporate the reverse sequence of the BCR exons 19, 20, and part of 22. NM_014549 contains a putative ORF of 428 aa. e, Partial-gene duplication of the last three exons of AK024854, a phorbolin-related gene, has lead to the formation of a five-exon fusion transcript. Exon 1 is derived from adjacent chromosome 22 sequence, whereas the terminal 4 exons are derived from the three duplicated exons of AK024854. Exon 2 and 3 correspond to exons 5 and 6 of AK024854. Exon 4 and 5 correspond to exon 7 of AK024854. f, Another partial gene duplication of a phorbolin-related transcript AF18240 (exons 1–4) has created a transcript represented at its 3′ end by EST AI092348. AI092348 has two exons with an ORF of at least 77 aa, extending in a 5′ orientation and terminating within the 3′ exon. The penultimate exon is derived from exon 2 of AF18240. g, Partial-gene duplication of the last three exons of crystallin beta B2 (CRYBB2) has lead to the formation of a new gene, represented by EST AW190323. Three exons of AF18240 are utilized in the new transcript, with the addition of two additional 5′ exons from the adjacent unduplicated sequence and a putatively new 3′ terminal exon from previously nonexonic sequence. The ESTs have ORFs ranging from 88–105 aa, compared to 205 aa for CRYBB2. _h,_ESTs supporting potential whole-gene duplication, with representative transcripts from both copies (AI669658 and AA228976). The most proximal transcript, AI669658, contains two exons with a predicted ORF of >90 aa. The distal transcript, AA228976, contains three exons with a predicted ORF of >59 aa. Both transcripts appear to extend in a 5′ orientation, with an undetermined intron-exon structure. The three transcripts not shown in this figure have been previously described: (1) multiple partial-gene duplications within the immunoglobulin lambda (IGL) locus (Kawasaki et al. 1997), and (2) whole-gene duplications of ret-like finger proteins (RLPF1, RLPF2, and RLPF3), creating two new genes with conserved intron-exon and coding structure (Seroussi et al. 1999).
Similar articles
- Conservation of pericentromeric duplications of a 200-kb part of the human 21q22.1 region in primates.
Orti R, Potier MC, Maunoury C, Prieur M, Créau N, Delabar JM. Orti R, et al. Cytogenet Cell Genet. 1998;83(3-4):262-5. doi: 10.1159/000015201. Cytogenet Cell Genet. 1998. PMID: 10072600 - Duplication of a gene-rich cluster between 16p11.1 and Xq28: a novel pericentromeric-directed mechanism for paralogous genome evolution.
Eichler EE, Lu F, Shen Y, Antonacci R, Jurecic V, Doggett NA, Moyzis RK, Baldini A, Gibbs RA, Nelson DL. Eichler EE, et al. Hum Mol Genet. 1996 Jul;5(7):899-912. doi: 10.1093/hmg/5.7.899. Hum Mol Genet. 1996. PMID: 8817324 - Evolutionary mechanisms shaping the genomic structure of the Williams-Beuren syndrome chromosomal region at human 7q11.23.
Antonell A, de Luis O, Domingo-Roura X, Pérez-Jurado LA. Antonell A, et al. Genome Res. 2005 Sep;15(9):1179-88. doi: 10.1101/gr.3944605. Genome Res. 2005. PMID: 16140988 Free PMC article. - Recent duplication, domain accretion and the dynamic mutation of the human genome.
Eichler EE. Eichler EE. Trends Genet. 2001 Nov;17(11):661-9. doi: 10.1016/s0168-9525(01)02492-1. Trends Genet. 2001. PMID: 11672867 Review.
Cited by
- TCEAL1 loss-of-function results in an X-linked dominant neurodevelopmental syndrome and drives the neurological disease trait in Xq22.2 deletions.
Hijazi H, Reis LM, Pehlivan D, Bernstein JA, Muriello M, Syverson E, Bonner D, Estiar MA, Gan-Or Z, Rouleau GA, Lyulcheva E, Greenhalgh L, Tessarech M, Colin E, Guichet A, Bonneau D, van Jaarsveld RH, Lachmeijer AMA, Ruaud L, Levy J, Tabet AC, Ploski R, Rydzanicz M, Kępczyński Ł, Połatyńska K, Li Y, Fatih JM, Marafi D, Rosenfeld JA, Coban-Akdemir Z, Bi W, Gibbs RA, Hobson GM, Hunter JV, Carvalho CMB, Posey JE, Semina EV, Lupski JR. Hijazi H, et al. Am J Hum Genet. 2022 Dec 1;109(12):2270-2282. doi: 10.1016/j.ajhg.2022.10.007. Epub 2022 Nov 10. Am J Hum Genet. 2022. PMID: 36368327 Free PMC article. - Structural divergence between the human and chimpanzee genomes.
Kehrer-Sawatzki H, Cooper DN. Kehrer-Sawatzki H, et al. Hum Genet. 2007 Feb;120(6):759-78. doi: 10.1007/s00439-006-0270-6. Epub 2006 Oct 26. Hum Genet. 2007. PMID: 17066299 Review. - Recurrent duplication-driven transposition of DNA during hominoid evolution.
Johnson ME; National Institute of Health Intramural Sequencing Center Comparative Sequencing Program; Cheng Z, Morrison VA, Scherer S, Ventura M, Gibbs RA, Green ED, Eichler EE. Johnson ME, et al. Proc Natl Acad Sci U S A. 2006 Nov 21;103(47):17626-31. doi: 10.1073/pnas.0605426103. Epub 2006 Nov 13. Proc Natl Acad Sci U S A. 2006. PMID: 17101969 Free PMC article. - Variance of IQ is partially dependent on deletion type among 1,427 22q11.2 deletion syndrome subjects.
Zhao Y, Guo T, Fiksinski A, Breetvelt E, McDonald-McGinn DM, Crowley TB, Diacou A, Schneider M, Eliez S, Swillen A, Breckpot J, Vermeesch J, Chow EWC, Gothelf D, Duijff S, Evers R, van Amelsvoort TA, van den Bree M, Owen M, Niarchou M, Bearden CE, Ornstein C, Pontillo M, Buzzanca A, Vicari S, Armando M, Murphy KC, Murphy C, Garcia-Minaur S, Philip N, Campbell L, Morey-Cañellas J, Raventos J, Rosell J, Heine-Suner D, Shprintzen RJ, Gur RE, Zackai E, Emanuel BS, Wang T, Kates WR, Bassett AS, Vorstman JAS, Morrow BE; International 22q11.2 Brain and Behavior Consortium. Zhao Y, et al. Am J Med Genet A. 2018 Oct;176(10):2172-2181. doi: 10.1002/ajmg.a.40359. Epub 2018 Oct 5. Am J Med Genet A. 2018. PMID: 30289625 Free PMC article. - Genomic structure and evolution of the ancestral chromosome fusion site in 2q13-2q14.1 and paralogous regions on other human chromosomes.
Fan Y, Linardopoulou E, Friedman C, Williams E, Trask BJ. Fan Y, et al. Genome Res. 2002 Nov;12(11):1651-62. doi: 10.1101/gr.337602. Genome Res. 2002. PMID: 12421751 Free PMC article.
References
Electronic-Database Information
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SMS [MIM <182290>], PWS [MIM <176270>], AS [MIM <105830>], NF1 [MIM <162200>], VCFS [MIM <192430>], DGS [MIM <188400>], CES [MIM <115470>], and CMT1A [MIM <118220>])
- RefSeq Database, http://www.ncbi.nlm.nih.gov/LocusLink/refseq.html (for accessions of the format NM_#####)
- RepeatMasker, http://repeatmasker.genome.washington.edu/
- Rocchi Lab Web site, http://www.biologia.uniba.it/22-paper/ (for all FISH images)
References
- Amos-Landgraf JM, Ji Y, Gottlieb W, Depinet T, Wandstrat AE, Cassidy SB, Driscoll DJ, Rogan PK, Schwartz S, Nicholls RD (1999) Chromosome breakage in the Prader-Willi and Angelman syndromes involves recombination between large, transcribed repeats at proximal and distal breakpoints. Am J Hum Genet 65:370–386 - PMC - PubMed
- Chen K, Manian P, Koeuth T, Potocki L, Zhao Q, Chinault A, Lee C, Lupski J (1997) Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat Genet 17:154–163 - PubMed
- Dorschner MO, Sybert VP, Weaver M, Pletcher BA, Stephens K (2000) NF1 microdeletion breakpoints are clustered at flanking repetitive sequences. Hum Mol Genet 9:35–46 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM058815/GM/NIGMS NIH HHS/United States
- GM58815/GM/NIGMS NIH HHS/United States
- E.0962/TI_/Telethon/Italy
- HG002385/HG/NHGRI NIH HHS/United States
- R01 HG002385/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous