Healing of burn wounds in transgenic mice overexpressing transforming growth factor-beta 1 in the epidermis - PubMed (original) (raw)
Healing of burn wounds in transgenic mice overexpressing transforming growth factor-beta 1 in the epidermis
L Yang et al. Am J Pathol. 2001 Dec.
Abstract
Transforming growth factor-beta (TGF-beta) isoforms are multifunctional cytokines that play an important role in wound healing. Transgenic mice overexpressing TGF-beta in the skin under control of epidermal-specific promoters have provided models to study the effects of increased TGF-beta on epidermal cell growth and cutaneous wound repair. To date, most of these studies used transgenic mice that overexpress active TGF-beta in the skin by modulating the latency-associated-peptide to prevent its association with active TGF-beta. The present study is the first to use transgenic mice that overexpress the natural form of latent TGF-beta 1 in the epidermis, driven by the keratin 14 gene promoter to investigate the effects of locally elevated TGF-beta 1 on the healing of partial-thickness burn wounds made on the back of the mice using a CO(2) laser. Using this model, we demonstrated activation of latent TGF-beta after wounding and determined the phenotypes of burn wound healing. We found that introduction of the latent TGF-beta1 gene into keratinocytes markedly increases the release and activation of TGF-beta after burn injury. Elevated local TGF-beta significantly inhibited wound re-epithelialization in heterozygous (42% closed versus 92% in controls, P < 0.05) and homozygous (25% versus 92%, P < 0.01) animals at day 12 after wounding. Interestingly, expression of type I collagen mRNA and hydroxyproline significantly increased in the wounds of transgenic mice, probably as a result of a paracrine effect of the transgene.
Figures
Figure 1.
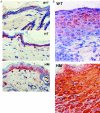
Immunolocalization of latent TGF-β1 in normal skin and wounds. Cryosections of the skin and wounds taken from transgenic and WT mice were immunostained with an antibody specific for human LAP1. A: In normal skin of WT mice, faint staining of LAP1 was seen in the epidermis and mainly extracellular in distribution. In the skin of HT and HM mice, high intensity of LAP staining is located mainly in the basal keratinocytes (arrows) and has a predominantly intracellular distribution. B: In day 6 wounds, latent TGF-β was observed throughout the epidermis adjacent to wound in transgenic mice, whereas, it was located mainly in cytoplasm of suprabasal keratinocytes in WT controls. Original magnifications, ×400.
Figure 2.
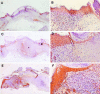
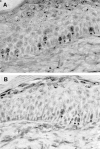
Immunolocalization of active TGF-β1 in the wounds. Wound sections taken at day 6 after wounding were immunostained with an antibody against active TGF-β1. In the wounds in WT mice (A and B), active TGF-β1 was present mainly in the suprabasal keratinocytes of the migrating epithelial sheet. The intensity of TGF-β staining was increased in wounds made in HT (C and D) and HM (E and F) mice and distributed throughout the epidermis. Note the wide epithelial gap in the wounds in transgenic mice. Original magnifications: ×20 (A, C, and E), ×100 (B, D, and F).
Figure 3.
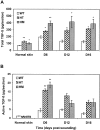
The levels of total and active TGF-β in the skin and wound sections. Cryosections of the skin and wounds taken at days 6, 12, and 16 after wounding from WT, HT, and HM mice were analyzed for total (A) and active (B) TGF-β using the PAI/L assay. The data represent mean values ± SEM of three separate experiments, each examined in triplicate with significant differences compared to controls at *P < 0.05 or **P < 0.01 (analysis of variance).
Figure 4.
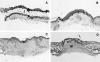
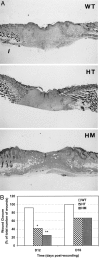
Histological features of laser wound healing in WT mice. Partial-thickness wounds were created on the back of the mice using a CO2 laser. Mice were sacrificed at different time points after wounding. Paraffin sections of the wounds were stained with H&E. A: Immediately after wounding the epidermis and dermis were damaged with a thick layer of fatty tissue remaining underneath. No immediate injury to the panniculus was observed. B: Day 6 wounds were characterized by infiltration of large number of inflammatory cells (arrow). The keratinocyte layers became thicker at the wound edge (arrowhead). The panniculus was no longer present at the wound sites. C: At day 12, re-epithelialization was complete with thick neo-epidermis at the wound surface. D: By day 16 the wound area became smaller and dense granulation tissue was seen in the wound. e, epidermis; f, fatty tissue; p, panniculus; gt, granulation tissue. Original magnifications, ×20.
Figure 5.
Retarded wound re-epithelialization in transgenic mice. A: Representative wound sections taken from WT, HT, and HM mice at day 12 after wounding illustrate delayed re-epithelialization in transgenic mice. B: Wound closure in WT, HT, and HM mice was assessed visually and histologically. Wounds were scored as open or closed. Forty-eight wounds were examined in each type of animal at 12 days and 16 days (Fisher’s exact test; *, P < 0.05; **, P < 0.01).
Figure 6.
Detection of proliferating cells in the epidermis of wounds by BrdU labeling. At day 6 after wounding, WT (A) and transgenic (B) mice were injected intraperitoneally with BrdU 1 hour before sacrifice. Wound sections were stained with an anti-BrdU antibody. BrdU-labeled nuclei are indicated by arrows. Original magnification, ×100.
Figure 7.
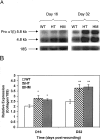
Expression of mRNA for type I collagen in the wounds. Total RNA was extracted from each wound taken from WT, HT, and HM mice at days 16 and 32, and type I procollagen mRNA was determined by Northern analysis. A: The autoradiogram shown is representative of three autoradiograms from separate experiments, demonstrating the pattern of 5.8-kb and 4.8-kb transcripts (top row) corresponding to pro α (I) procollagen mRNA. The bottom row shows the profile of the 18S ribosomal RNA used as a control of RNA loading, obtained by rehybridization of the blot shown in the top row. B: Quantitative comparison of procollagen mRNA expression in WT, HT, and HM wounds. The relative intensity of procollagen mRNA to 18S ribosomal RNA was calculated and shown as the mean ± SEM with significant differences at *P < 0.05 or **P < 0.01 (analysis of variance).
Figure 8.
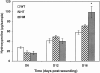
The levels of hydroxyproline in wound tissue. Wound samples taken from WT, HT, and HM mice at different time points were processed for mass spectrometric analysis for 4-hydroxyproline. The data represent mean values ± SEM of four separate experiments with a significant difference at *P < 0.05 (analysis of variance).
Similar articles
- Development, characterization, and wound healing of the keratin 14 promoted transforming growth factor-beta1 transgenic mouse.
Chan T, Ghahary A, Demare J, Yang L, Iwashina T, Scott PG, Tredget EE. Chan T, et al. Wound Repair Regen. 2002 May-Jun;10(3):177-87. doi: 10.1046/j.1524-475x.2002.11101.x. Wound Repair Regen. 2002. PMID: 12100379 - Transforming growth factor-beta(1), -beta(2), -beta(3), basic fibroblast growth factor and vascular endothelial growth factor expression in keratinocytes of burn scars.
Hakvoort T, Altun V, van Zuijlen PP, de Boer WI, van Schadewij WA, van der Kwast TH. Hakvoort T, et al. Eur Cytokine Netw. 2000 Jun;11(2):233-39. Eur Cytokine Netw. 2000. PMID: 10903802 - Suppression of keratin 15 expression by transforming growth factor beta in vitro and by cutaneous injury in vivo.
Werner S, Munz B. Werner S, et al. Exp Cell Res. 2000 Jan 10;254(1):80-90. doi: 10.1006/excr.1999.4726. Exp Cell Res. 2000. PMID: 10623468 - Role of TGF beta-mediated inflammation in cutaneous wound healing.
Wang XJ, Han G, Owens P, Siddiqui Y, Li AG. Wang XJ, et al. J Investig Dermatol Symp Proc. 2006 Sep;11(1):112-7. doi: 10.1038/sj.jidsymp.5650004. J Investig Dermatol Symp Proc. 2006. PMID: 17069018 Review. - TGF beta regulation of cell proliferation.
Moses HL, Arteaga CL, Alexandrow MG, Dagnino L, Kawabata M, Pierce DF Jr, Serra R. Moses HL, et al. Princess Takamatsu Symp. 1994;24:250-63. Princess Takamatsu Symp. 1994. PMID: 8983080 Review.
Cited by
- Mechanical and Immunological Regulation in Wound Healing and Skin Reconstruction.
Kimura S, Tsuji T. Kimura S, et al. Int J Mol Sci. 2021 May 22;22(11):5474. doi: 10.3390/ijms22115474. Int J Mol Sci. 2021. PMID: 34067386 Free PMC article. Review. - Role of TGF-β in Skin Chronic Wounds: A Keratinocyte Perspective.
Liarte S, Bernabé-García Á, Nicolás FJ. Liarte S, et al. Cells. 2020 Jan 28;9(2):306. doi: 10.3390/cells9020306. Cells. 2020. PMID: 32012802 Free PMC article. Review. - Smad4 disruption accelerates keratinocyte reepithelialization in murine cutaneous wound repair.
Yang L, Li W, Wang S, Wang L, Li Y, Yang X, Peng R. Yang L, et al. Histochem Cell Biol. 2012 Oct;138(4):573-82. doi: 10.1007/s00418-012-0974-8. Epub 2012 May 30. Histochem Cell Biol. 2012. PMID: 22644379 - Role of Mitogen-Activated Protein Kinases in the Formation of Hypertrophic Scar with Model of Lipopolysaccharide Stimulated Skin Fibroblast Cells.
Wang W, Li G, Yang H. Wang W, et al. Pak J Med Sci. 2018 Jan-Feb;34(1):215-220. doi: 10.12669/pjms.341.13636. Pak J Med Sci. 2018. PMID: 29643910 Free PMC article. - Dermal Fibroblasts from the Red Duroc Pig Have an Inherently Fibrogenic Phenotype: An In Vitro Model of Fibroproliferative Scarring.
Sood RF, Muffley LA, Seaton ME, Ga M, Sirimahachaiyakul P, Hocking AM, Gibran NS. Sood RF, et al. Plast Reconstr Surg. 2015 Nov;136(5):990-1000. doi: 10.1097/PRS.0000000000001704. Plast Reconstr Surg. 2015. PMID: 26505702 Free PMC article.
References
- Roberts AB, Sporn MB: Transforming growth factor-β. Clark RAF eds. The Molecular and Cellular Biology of Wound Repair. 1996, :pp 275-308 Plenum Press, New York
- O’Kane S, Ferguson MWJ: Transforming growth factor βs and wound healing. Int J Biochem Cell Biol 1997, 29:63-78 - PubMed
- Massagué J: The transforming growth factor-β family. Annu Rev Cell Biol 1990, 6:597-641 - PubMed
- Roberts AB, Sporn MB: The transforming growth factor-βs. Sporn MB Roberts AB eds. Peptide Growth Factors and Their Receptors. 1990, :pp 419-472 Springer, Berlin
- Miyazono K, Ichijo H, Heldin C-H: Transforming growth factor-β: latent forms, binding proteins and receptors. Growth Factors 1993, 8:11-22 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous