The septin CDCrel-1 is dispensable for normal development and neurotransmitter release - PubMed (original) (raw)
The septin CDCrel-1 is dispensable for normal development and neurotransmitter release
Xiao-Rong Peng et al. Mol Cell Biol. 2002 Jan.
Abstract
Septins are GTPases required for the completion of cytokinesis in a variety of organisms, yet their role in this process is not known. Septins may have additional functions since the mammalian septin CDCrel-1 is predominantly expressed in the nervous system, a largely postmitotic tissue. While relatively little is known about the function of this protein, we have previously shown that it is involved in regulated secretion. In addition, the gene encoding this protein maps to a locus often deleted in velo-cardiofacial and DiGeorge syndromes, and CDCrel-1 has recently been shown to be a direct target of the E3 ubiquitin ligase activity of Parkin, a causative agent in autosomal recessive forms of Parkinson's disease. Here we show that CDCrel-1 expression rises at the time of synaptic maturation and that CDCrel-1 is present in a complex that includes the septins Nedd5 and CDC10. To investigate its function in the nervous system, we generated homozygotic CDCrel-1 null mice and showed that these mice appear normal with respect to synaptic properties and hippocampal neuron growth in vitro. Moreover, we found that while the expression of a number of synaptic proteins is not affected in the CDCrel-1 mutant mice, the expression of other septins is altered. Together, these data suggest that CDCrel-1 is not essential for neuronal development or function, and that changes in expression of other septins may account for its functional redundancy.
Figures
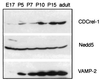
FIG. 1.
Developmental expression of CDCrel-1 in the mouse brain. Brains were isolated from embryonic (embryonic day 17 [E17]) and postnatal (postnatal day 5 [P5], P7, P10, P15, and adult) mice. Total homogenates were prepared for each, and 40 μg of protein was electrophoresed and blotted with antibodies specific to the proteins indicated at the right.
FIG. 2.
CDCrel-1 coimmunoprecipitates with other brain septins. The P2 fraction from a mouse brain homogenate (18) was prepared and solubilized with _n_-octylglucoside, and then it was centrifuged to separate the soluble and insoluble proteins (left side of blot). The soluble fraction was then used as the starting material for immunoprecipitations with the antibodies indicated above the right portion of the blots. Precipitates were electrophoresed under nonreducing conditions and probed with the antibodies indicated at the right. Control immunoglobulin G (IgG) was nonspecific rabbit serum. Reactivity to the free heavy chain was observed in the anti-CDC10 immunoprecipitation lanes (bottom two blots) and in the IgG lane (bottom blot) because these were crude sera, but these were located above the septin bands. No reactivity was seen on the top blot since the blotting antibody was from a mouse. Of the _n_-octylglucoside soluble material used for the immunoprecipitations, 2% was loaded in the first lane.
FIG. 3.
Generation of CDCrel-1-deficient mice by homologous recombination. (A) Construction of targeting vector. Schematic representation of the CDCrel-1 protein (top) and the wild-type genomic locus, showing the targeting vector and targeted genomic locus. The location of the probes used for Southern blotting to identify targeting events are shown below the protein. Exons are indicated by grey boxes, and exons 1 to 3 are not shown. The restriction enzymes indicated are _Bgl_II (Bgl), _Eco_RI (R1), _Hin_dIII (H3), _Xho_I (Xho), and _Sal_I (Sal). (B) Southern blots of DNA extracted from mouse tails. Genomic DNA digested with _Bgl_II and probed with the 5′ probe reveals the presence of the neo cassette replacing exons 4 to 6. The _Bgl_II fragment on the wild-type CDCrel-1 locus is approximately 16.5 kb while the 5′ _Bgl_II fragment of the targeted CDCrel-1 locus is approximately 9.5 kb. (C) Mutant mice lack the CDCrel-1 protein. Western blots show ∼10 μg of protein extracted from total brain electrophoresed and probed with anti-CDCrel-1 monoclonal antibody. Chimeras were generated from two independent ES clones (4-2G and 4-4B), and the progeny of both were tested. A reduction in CDCrel-1 levels is apparent in heterozygotes, and the signal is absent from homozygous mutant mice even after prolonged exposure.
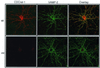
FIG. 4.
Hippocampal neurons from CDCrel-1-deficient mice appear normal in vitro. Primary cultures were prepared from the hippocampi of CDCrel-1-deficient mice (mt) and their wild-type littermates (wt). After 11 days in vitro, the cells were fixed and stained with antibodies against CDCrel-1 (red) and VAMP-2 (green). Confocal images were collected and shown. CDCrel-1-deficient neurons had only background labeling due to the secondary antibody. The merged images are shown in the overlay column.
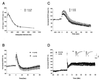
FIG. 5.
Electrophysiology of hippocampal slices from CDCrel-1-deficient mice. (A) Normal paired-pulse facilitation in CDCrel-1-deficient mice. The plot summarizes facilitation of the second fEPSP slope compared to the first one as a function of the interpulse interval. (B) Normal posttetanic potentiation in CDCrel-1-deficient mice. Brief (1 s) 100-Hz stimulation was given at the zero time point in the presence of 50 μM (−)-2-amino-5-phosphonopentanoic acid, and the fEPSPs were recorded immediately after the tetanus. (C) Synaptic depression in response to sustained synaptic activities in the knockout mice. Repetitive stimuli (5 Hz, lasting 120 s) were applied at the zero time point, and the fEPSP to each stimulus was recorded. Each data point represents the averaged slope of 20 responses (2 s). (D) Hippocampal CA1 LTP. LTP was induced by tetanic stimulation consisting of 2 trains of 100 Hz lasting 1 s, at an intertrain interval of 10 s, delivered at the zero time point. Above the plot are the representative traces (average of 4 sweeps) of fEPSP obtained immediately before (trace 1) and 50 min after (trace 2) the tetanic stimulation.
FIG. 6.
Altered expression of septins, but not other synaptic proteins, in the brains of CDCrel-1-deficient mice. Wild-type and CDCrel-1-deficient brains were fractionated according to the procedure of Huttner et al. (18), and 20 μg of protein from each fraction was loaded onto each lane. Lanes: H, total homogenate; S1, low-speed supernatant; P2, crude synaptosomal fraction; P3, high-speed pellet of synaptosome-depleted sample; S3, cytosolic high-speed supernatant; LP1, low-speed pellet of lysed synaptosome containing membrane fractions; and LP2, high-speed pellet of lysed synaptosomes containing synaptic vesicles. Blots were probed with the antibodies indicated at the right.
Similar articles
- The septin CDCrel-1 binds syntaxin and inhibits exocytosis.
Beites CL, Xie H, Bowser R, Trimble WS. Beites CL, et al. Nat Neurosci. 1999 May;2(5):434-9. doi: 10.1038/8100. Nat Neurosci. 1999. PMID: 10321247 - Human septin-septin interaction: CDCrel-1 partners with KIAA0202.
Bläser S, Jersch K, Hainmann I, Wunderle D, Zgaga-Griesz A, Busse A, Zieger B. Bläser S, et al. FEBS Lett. 2002 May 22;519(1-3):169-72. doi: 10.1016/s0014-5793(02)02749-7. FEBS Lett. 2002. PMID: 12023038 - Superfluous role of mammalian septins 3 and 5 in neuronal development and synaptic transmission.
Tsang CW, Fedchyshyn M, Harrison J, Xie H, Xue J, Robinson PJ, Wang LY, Trimble WS. Tsang CW, et al. Mol Cell Biol. 2008 Dec;28(23):7012-29. doi: 10.1128/MCB.00035-08. Epub 2008 Sep 22. Mol Cell Biol. 2008. PMID: 18809578 Free PMC article. - Mammalian septin function in hemostasis and beyond.
Martinez C, Ware J. Martinez C, et al. Exp Biol Med (Maywood). 2004 Dec;229(11):1111-9. doi: 10.1177/153537020422901105. Exp Biol Med (Maywood). 2004. PMID: 15564437 Review. - Septin localization across kingdoms: three themes with variations.
Lindsey R, Momany M. Lindsey R, et al. Curr Opin Microbiol. 2006 Dec;9(6):559-65. doi: 10.1016/j.mib.2006.10.009. Epub 2006 Oct 25. Curr Opin Microbiol. 2006. PMID: 17067846 Review.
Cited by
- Septins, a cytoskeletal protein family, with emerging role in striated muscle.
Gönczi M, Dienes B, Dobrosi N, Fodor J, Balogh N, Oláh T, Csernoch L. Gönczi M, et al. J Muscle Res Cell Motil. 2021 Jun;42(2):251-265. doi: 10.1007/s10974-020-09573-8. Epub 2020 Jan 18. J Muscle Res Cell Motil. 2021. PMID: 31955380 Free PMC article. Review. - Alterations of social interaction through genetic and environmental manipulation of the 22q11.2 gene Sept5 in the mouse brain.
Harper KM, Hiramoto T, Tanigaki K, Kang G, Suzuki G, Trimble W, Hiroi N. Harper KM, et al. Hum Mol Genet. 2012 Aug 1;21(15):3489-99. doi: 10.1093/hmg/dds180. Epub 2012 May 15. Hum Mol Genet. 2012. PMID: 22589251 Free PMC article. - Septin 4, the drosophila ortholog of human CDCrel-1, accumulates in parkin mutant brains and is functionally related to the Nedd4 E3 ubiquitin ligase.
Muñoz-Soriano V, Nieto-Arellano R, Paricio N. Muñoz-Soriano V, et al. J Mol Neurosci. 2012 Sep;48(1):136-43. doi: 10.1007/s12031-012-9788-3. Epub 2012 May 6. J Mol Neurosci. 2012. PMID: 22562816 - The role of septin 7 in physiology and pathological disease: A systematic review of current status.
Wang X, Fei F, Qu J, Li C, Li Y, Zhang S. Wang X, et al. J Cell Mol Med. 2018 Jul;22(7):3298-3307. doi: 10.1111/jcmm.13623. Epub 2018 Mar 30. J Cell Mol Med. 2018. PMID: 29602250 Free PMC article. - Production and analysis of a mammalian septin hetero-octamer complex.
DeRose BT, Kelley RS, Ravi R, Kokona B, Beld J, Spiliotis ET, Padrick SB. DeRose BT, et al. Cytoskeleton (Hoboken). 2020 Nov;77(11):485-499. doi: 10.1002/cm.21643. Epub 2020 Nov 23. Cytoskeleton (Hoboken). 2020. PMID: 33185030 Free PMC article.
References
- Beites, C. L., H. Xie, R. Bowser, and W. S. Trimble. 1999. The septin CDCrel-1 binds syntaxin and inhibits exocytosis. Nat. Neurosci. 2:434–439. - PubMed
- Byers, B., and L. Goetsch. 1976. Loss of the filamentous ring in cytokinesis-defective mutants of budding yeast. J. Cell Biol. 70:35.
- Caltagarone, J., J. Rhodes, W. G. Honer, and R. Bowser. 1998. Localization of a novel septin protein, hCDCrel-1, in neurons of human brain. Neuroreport 9:2907–2912. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous