Golgi-to-endoplasmic reticulum (ER) retrograde traffic in yeast requires Dsl1p, a component of the ER target site that interacts with a COPI coat subunit - PubMed (original) (raw)
Golgi-to-endoplasmic reticulum (ER) retrograde traffic in yeast requires Dsl1p, a component of the ER target site that interacts with a COPI coat subunit
B A Reilly et al. Mol Biol Cell. 2001 Dec.
Free PMC article
Abstract
DSL1 was identified through its genetic interaction with SLY1, which encodes a t-SNARE-interacting protein that functions in endoplasmic reticulum (ER)-to-Golgi traffic. Conditional dsl1 mutants exhibit a block in ER-to-Golgi traffic at the restrictive temperature. Here, we show that dsl1 mutants are defective for retrograde Golgi-to-ER traffic, even under conditions where no anterograde transport block is evident. These results suggest that the primary function of Dsl1p may be in retrograde traffic, and that retrograde defects can lead to secondary defects in anterograde traffic. Dsl1p is an ER-localized peripheral membrane protein that can be extracted from the membrane in a multiprotein complex. Immunoisolation of the complex yielded Dsl1p and proteins of approximately 80 and approximately 55 kDa. The approximately 80-kDa protein has been identified as Tip20p, a protein that others have shown to exist in a tight complex with Sec20p, which is approximately 50 kDa. Both Sec20p and Tip20p function in retrograde Golgi-to-ER traffic, are ER-localized, and bind to the ER t-SNARE Ufe1p. These findings suggest that an ER-localized complex of Dsl1p, Sec20p, and Tip20p functions in retrograde traffic, perhaps upstream of a Sly1p/Ufe1p complex. Last, we show that Dsl1p interacts with the delta-subunit of the retrograde COPI coat, Ret2p, and discuss possible roles for this interaction.
Figures
Figure 1
dsl1-7 shows a CPY trafficking block in the absence of an invertase block. Log phase wild-type (RSY255), dsl1-4 (GWY230), and dsl1-7 (GWY233) strains were grown on media lacking methionine and containing 2% raffinose and 0.1% glucose for 1 h before a shift to the indicated temperature for 20 min. Strains were labeled with 35S-amino acids for 10 min, chased for 20 min at the same temperature, and lysed. CPY (top) or invertase (bottom) was then immunoprecipitated from each sample, separated by SDS-7% PAGE, and visualized by autoradiography. The migration of the pre-Golgi (p1), Golgi (p2), and mature (m) forms of CPY, and of the ER and Golgi forms of invertase is indicated on the right. Immunoprecipitations from lysates of sec17-1 (GWY270) and _pep4_Δ (MS1554) strains incubated at 37°C were used as controls for the p1 and p2 forms of CPY, and the ER and mature form of invertase, respectively. INV, invertase; IP, immunoprecipitation.
Figure 2
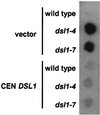
dsl1ts strains secrete ER resident Kar2p. Log phase wild-type (RSY255), dsl1-4 (GWY230), and dsl1-7 (GWY233) strains bearing pRS416 (vector) or pSC2 (CEN DSL1) were spotted onto rich medium, overlayed with nitrocellulose, and allowed to grow at 37°C for 16 h. Kar2p that had been secreted and bound to the nitrocellulose was detected by immunoblotting with α-Kar2p antibody and ECL PLUS detection.
Figure 3
Retrograde transport is inhibited in dsl1 mutants. MATa Ura+ Lys− wild-type (PC13), ret1-1 (PC75), dsl1-4 (GWY379), dsl1-7 (GWY380) strains were grown on nonselective medium for 2 h at 23, 27, 30, 34, or 37°C and then replica plated to a lawn of wild-type cells of the opposite mating type (α Ura−Lys+) and incubated at the same temperature for 6 h. Mating was then analyzed by growth on medium selective for diploids (SC-Ura-Lys). Note that although 30°C is a restrictive temperature for the dsl1-7 strain, mating is detected at this temperature. Perhaps the cells die slowly at this temperature, giving them an opportunity to mate and then survive as heterozygous diploids.
Figure 4
dsl1-7 exhibits synthetic lethal interactions with both sec20-1 and tip20-5. dsl1-7 (GWY414) was mated to sec20-1 (RSY275) (A) or tip20-5 (PC137) (B), and the resulting diploid strains were sporulated, dissected onto YPD plates, and incubated at 23°C for 2 d. Spores were then replica plated to YPD and incubated at the indicated temperatures for 2 d. Two representative tetratypes are shown. Genotypes (indicated on the right; D, DSL1; d, dsl1-7; S, SEC20, s, sec20-1; T, TIP20; t, tip20-5) were determined by complementation of the temperature sensitivity with plasmid-borne SEC20, TIP20, and/or DSL1.
Figure 5
Dsl1p is a peripheral membrane protein. (A) Characterization of the anti-Dsl1p antibody. Total yeast lysates prepared from a wild-type strain (RSY255) and an identical strain expressing DSL1 from a high-copy plasmid (pSC10) (left) or a wild-type strain (KRY8) and the dsl1-1 strain (GWY167) containing pSV12 (right) were separated by SDS-7% PAGE and immunoblotted with antibodies against Dsl1p. The migration of molecular mass standards (New England Biolabs, Beverly, MA) is shown on the left. Note that the dsl1-1 strain harbors the pSV12 _SLY1-20_-bearing plasmid because it suppresses the inviability of the dsl1-1 mutation (VanRheenen et al., 2001). (B) Subcellular fractionation of Dsl1p. A yeast lysate was centrifuged at 175,000 × g and separated into supernatant (S) and pellet (P) fractionation, which were resolved by SDS-12% PAGE and immunoblotted with antibodies against Dsl1p, Sed5p, Sec61p, or PGK, as indicated. (C) Dsl1p is a tightly bound peripheral membrane protein. Total yeast membranes were isolated on a step gradient and incubated for 45 min in either buffer, 1% Triton X-100, 1 M NaCl, or 100 mM Na2CO3, pH 11. Reaction mixtures were separated into supernatant (S) and pellet (P) fractions by centrifugation at 175,000 × g for 60 min and analyzed by SDS-12% PAGE and immunoblotting with antibodies against Dsl1p (top) or Sed5p (bottom).
Figure 6
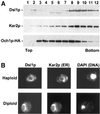
Dsl1p is an ER protein. (A) Dsl1p comigrates with Kar2p and not with Och1p-HA by buoyant density centrifugation. Membranes from an Och1p-HA-expressing strain (RSY255 bearing pSV66) were separated by buoyant density centrifugation. Gradient fractions were analyzed by immunoblotting with anti-Dsl1p, anti-Kar2p, and anti-HA antibodies as indicated. (B) Dsl1p localizes to perinuclear regions similar to Kar2p. Haploid or diploid strains expressing Dsl1p-HA (pSV61) were examined by indirect immunofluorescence for Dsl1p and Kar2p, as indicated. Diamidophenylindole (DAPI) staining of the nucleus is shown on the right.
Figure 7
Dsl1p is present in a large complex that contains Tip20p. (A) Dsl1p is part of a large complex. Size exclusion chromatography of Triton X-100H- or n-dodecyl-β-
d
-maltoside-extracted membranes was followed by immunoblotting fractions with anti-Dsl1p. The asterisks indicate a potential proteolytic product generated during chromatography. L represents a sample of the material loaded on to the column. (B) Dsl1p interacts with proteins of ∼80 and ∼50 kDa. Triton X-100H-solubilized membranes from a strain expressing Dsl1p-HA (pSV61, left lane) or wild-type Dsl1p (pSV67, right lane) were immunoprecipitated with anti-HA, and the harvested proteins were subjected to SDS-12% PAGE and silver staining. Proteolytic fragments of Dsl1p are indicated with an asterisk. (C) Dsl1p coimmunoprecipitates with Tip20p-myc. Lysate from a log phase wild-type strain (RSY255) bearing TIP20-myc (pTM2) was incubated in the presence of 1% Triton X-100. Precleared supernatant (Total, lane 1) was subjected to immunoprecipitation with anti-myc antibody (9E10)-conjugated protein A-Sepharose beads in the presence of buffer (lanes 2 and 3) or 1 μg/ml myc peptide (lanes 4 and 5). The immunoprecipitated (IP) and nonbound protein (S) fractions were probed for Dsl1p and Sec61p by immunoblotting followed by ECL PLUS detection.
Figure 8
Dsl1p interacts with Ret2p/δ-COP in a two-hybrid system. (A) Log phase two-hybrid strain (PJ69–4A) bearing bait (DNA-binding domain) and prey (activation domain) vectors as indicated were serially diluted and spotted onto YPD medium to assess viability, or onto synthetic complete medium lacking adenine to assay the two-hybrid interaction. Coexpression of p53 fused to the Gal4p-BD and T-antigen fused to the Gal4p-AD was used as a positive control. (B) Dsl1p-interacting domain of Ret2p. The RET2 open reading frame is 1641 base pairs in length. RET2 isolate 1 contains a library insert corresponding to bases 422-1196 and RET2 isolates 2 and 3 start at bases 466 and 362, respectively, and continue past the stop codon. Thus, the central portion (bases 466-1196) of Ret2p encodes a Dsl1p-interacting region.
Similar articles
- Sec20p-interacting proteins (Tip20p, Ufe1p) in the retrograde secretory pathway of the fungal pathogen Candida albicans.
Weber Y, Swoboda RK, Ernst JF. Weber Y, et al. Mol Genet Genomics. 2002 Dec;268(4):468-76. doi: 10.1007/s00438-002-0777-z. Epub 2002 Nov 16. Mol Genet Genomics. 2002. PMID: 12471444 - Dsl1p, Tip20p, and the novel Dsl3(Sec39) protein are required for the stability of the Q/t-SNARE complex at the endoplasmic reticulum in yeast.
Kraynack BA, Chan A, Rosenthal E, Essid M, Umansky B, Waters MG, Schmitt HD. Kraynack BA, et al. Mol Biol Cell. 2005 Sep;16(9):3963-77. doi: 10.1091/mbc.e05-01-0056. Epub 2005 Jun 15. Mol Biol Cell. 2005. PMID: 15958492 Free PMC article. - Dsl1p/Zw10: common mechanisms behind tethering vesicles and microtubules.
Schmitt HD. Schmitt HD. Trends Cell Biol. 2010 May;20(5):257-68. doi: 10.1016/j.tcb.2010.02.001. Epub 2010 Mar 11. Trends Cell Biol. 2010. PMID: 20226673 Review. - Exiting the endoplasmic reticulum.
Gorelick FS, Shugrue C. Gorelick FS, et al. Mol Cell Endocrinol. 2001 May 25;177(1-2):13-8. doi: 10.1016/s0303-7207(01)00438-5. Mol Cell Endocrinol. 2001. PMID: 11377815 Review.
Cited by
- Activation of Gαi at the Golgi by GIV/Girdin imposes finiteness in Arf1 signaling.
Lo IC, Gupta V, Midde KK, Taupin V, Lopez-Sanchez I, Kufareva I, Abagyan R, Randazzo PA, Farquhar MG, Ghosh P. Lo IC, et al. Dev Cell. 2015 Apr 20;33(2):189-203. doi: 10.1016/j.devcel.2015.02.009. Epub 2015 Apr 9. Dev Cell. 2015. PMID: 25865347 Free PMC article. - Moonlighting functions of the NRZ (mammalian Dsl1) complex.
Tagaya M, Arasaki K, Inoue H, Kimura H. Tagaya M, et al. Front Cell Dev Biol. 2014 Jun 11;2:25. doi: 10.3389/fcell.2014.00025. eCollection 2014. Front Cell Dev Biol. 2014. PMID: 25364732 Free PMC article. Review. - Multipronged interaction of the COG complex with intracellular membranes.
Willett R, Pokrovskaya I, Kudlyk T, Lupashin V. Willett R, et al. Cell Logist. 2014 Jan 1;4(1):e27888. doi: 10.4161/cl.27888. Epub 2014 Feb 13. Cell Logist. 2014. PMID: 24649395 Free PMC article. - Coat-tether interaction in Golgi organization.
Guo Y, Punj V, Sengupta D, Linstedt AD. Guo Y, et al. Mol Biol Cell. 2008 Jul;19(7):2830-43. doi: 10.1091/mbc.e07-12-1236. Epub 2008 Apr 23. Mol Biol Cell. 2008. PMID: 18434597 Free PMC article. - Membrane detachment is not essential for COG complex function.
Climer LK, Pokrovskaya ID, Blackburn JB, Lupashin VV. Climer LK, et al. Mol Biol Cell. 2018 Apr 15;29(8):964-974. doi: 10.1091/mbc.E17-11-0694. Epub 2018 Mar 30. Mol Biol Cell. 2018. PMID: 29467253 Free PMC article.
References
- Adams A, Gottschling DE, Kaiser DE, Stearns T. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Laboratory Press; 1998.
- Andag U, Neuman T, Schmitt HD. The coatomer interacting protein is required for Golgi-to-ER retrieval in yeast. J Biol Chem. 2001;276:39150–39160. - PubMed
- Ballensiefen W, Ossipov D, Schmitt HD. Recycling of the yeast v-SNARE Sec22p involves COPI-proteins and the ER transmembrane proteins Ufe1p and Sec20p. J Cell Sci. 1998;111:1507–1520. - PubMed
- Barlowe C. Traffic COPs of the early secretory pathway. Traffic. 2000;1:371–377. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases