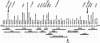A mutation in the LDL receptor-related protein 5 gene results in the autosomal dominant high-bone-mass trait - PubMed (original) (raw)
doi: 10.1086/338450. Epub 2001 Dec 3.
John P Carulli, Richard G Del Mastro, Josée Dupuis, Mark Osborne, Colleen Folz, Susan P Manning, Pamela M Swain, Shan-Chuan Zhao, Brenda Eustace, Michelle M Lappe, Lia Spitzer, Susan Zweier, Karen Braunschweiger, Youssef Benchekroun, Xintong Hu, Ronald Adair, Linda Chee, Michael G FitzGerald, Craig Tulig, Anthony Caruso, Nia Tzellas, Alicia Bawa, Barbara Franklin, Shannon McGuire, Xavier Nogues, Gordon Gong, Kristina M Allen, Anthony Anisowicz, Arturo J Morales, Peter T Lomedico, Susan M Recker, Paul Van Eerdewegh, Robert R Recker, Mark L Johnson
Affiliations
- PMID: 11741193
- PMCID: PMC419982
- DOI: 10.1086/338450
A mutation in the LDL receptor-related protein 5 gene results in the autosomal dominant high-bone-mass trait
Randall D Little et al. Am J Hum Genet. 2002 Jan.
Abstract
Osteoporosis is a complex disease that affects >10 million people in the United States and results in 1.5 million fractures annually. In addition, the high prevalence of osteopenia (low bone mass) in the general population places a large number of people at risk for developing the disease. In an effort to identify genetic factors influencing bone density, we characterized a family that includes individuals who possess exceptionally dense bones but are otherwise phenotypically normal. This high-bone-mass trait (HBM) was originally localized by linkage analysis to chromosome 11q12-13. We refined the interval by extending the pedigree and genotyping additional markers. A systematic search for mutations that segregated with the HBM phenotype uncovered an amino acid change, in a predicted beta-propeller module of the low-density lipoprotein receptor-related protein 5 (LRP5), that results in the HBM phenotype. During analysis of >1,000 individuals, this mutation was observed only in affected individuals from the HBM kindred. By use of in situ hybridization to rat tibia, expression of LRP5 was detected in areas of bone involved in remodeling. Our findings suggest that the HBM mutation confers a unique osteogenic activity in bone remodeling, and this understanding may facilitate the development of novel therapies for the treatment of osteoporosis.
Figures
Figure 1
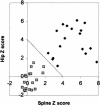
BMD measurements. A plot of the hip versus spine BMD measurements of the family with HBM is shown. Blackened diamonds indicate individuals with the mutation, and shaded squares indicate individuals without the mutation. The criterion used for assigning affection status was a sum >4 of Z scores from hip and spine, as indicated by the diagonal line.
Figure 2
HBM pedigree and haplotypes of the individuals used in the genetic–linkage studies. Blackened symbols represent affected individuals. Symbols containing “N” indicate unaffected individuals. Numbers beneath the symbols show the identification numbers, the Z scores for the spinal and hip BMD, and the alleles for the critical markers on chromosome 11. Question marks (?) denote unknown affection status, genotype, or phase. Untyped genotypes inferred with certainty are included in parentheses. The striped haplotype is shared by all affected individuals; critical crossovers were identified in individuals 44 and 46. Asterisks (*) indicate the 16 additional individuals used to narrow the region. Arrow indicates the proband.
Figure 3
Physical map of the HBM interval. STS markers derived from genes, expressed-sequence tags, microsatellites, random sequences, and BAC end sequences are denoted above the long horizontal line. STSs derived from BAC end sequences are listed with the BAC library address followed by L or R, for the left or right end of the clone, respectively; end sequences derived from BACs from the Genome Systems library are indicated with GS. The two large arrows indicate the location of genetic markers that define the HBM critical region. The horizontal lines below the STSs indicate a minimal tiling path of BAC clones that were sequenced. Open circles indicate that the marker did not amplify from the corresponding BAC. All clones were from the CITB library except for the two preceded by an RPCI-11 prefix. A region that was underrepresented in the library is indicated as a gap. Genes that were analyzed for mutations are shown in their approximate location at the top of the figure. Genes with a prefix of Hs correspond to UniGene entries.
Figure 4
HBM mutation and domain structure of the LRP5 protein. A, Sequence traces illustrating the HBM mutation (arrow) for a core four-member family. Identification numbers are as in figure 2. B, Domain organization of LRP5. The site of the glycine-to-valine change that occurs in the HBM protein is indicated with an arrow. C, Similarity of LRP5 and LDLR. A ribbon representation of LDLR, in which colors are based on homology with the first propeller (and EGF) domains of LRP5, is shown as viewed from the top. Identities are shown in red, and similarities are shown in pink. The location of G516 of the mature LDLR protein (comparable to residue G171 in LRP5) is shown in green. D, Simulation of the LRTP5 G171V mutation in the LDLR YWTD-EGF domain pair. The glycine at amino acid 516 of the mature LDLR protein was replaced with a valine, and the resulting three-dimensional structures were superimposed and viewed from the side. The wild-type structure is shown in blue with G516 in green, and the G516V mutant structure is shown in white with the V516 residue in red.
Figure 5
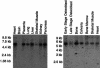
Northern blot analyses showing the expression of LRP5 in various tissues.
Figure 6
In situ hybridization of the LRP5 gene in bone tissue. A, Localization of LRP5 transcripts in rat endosteum by in situ hybridization with antisense and sense probes; original magnification ×400. Arrow indicates bone-lining cells. B, Localization of LRP5 transcripts in rat metaphysis and growth plate by in situ hybridization with antisense and sense probes; original magnification ×100. Boxes enclose regions that are shown at higher magnification in panel C. C, Localization of LRP5 transcripts in rat proximal metaphysis by in situ hybridization with antisense and sense probes; original magnification ×400.
Similar articles
- New explanation for autosomal dominant high bone mass: Mutation of low-density lipoprotein receptor-related protein 6.
Whyte MP, McAlister WH, Zhang F, Bijanki VN, Nenninger A, Gottesman GS, Lin EL, Huskey M, Duan S, Dahir K, Mumm S. Whyte MP, et al. Bone. 2019 Oct;127:228-243. doi: 10.1016/j.bone.2019.05.003. Epub 2019 May 11. Bone. 2019. PMID: 31085352 - Oropharyngeal skeletal disease accompanying high bone mass and novel LRP5 mutation.
Rickels MR, Zhang X, Mumm S, Whyte MP. Rickels MR, et al. J Bone Miner Res. 2005 May;20(5):878-85. doi: 10.1359/JBMR.041223. Epub 2004 Dec 20. J Bone Miner Res. 2005. PMID: 15824861 - High bone density due to a mutation in LDL-receptor-related protein 5.
Boyden LM, Mao J, Belsky J, Mitzner L, Farhi A, Mitnick MA, Wu D, Insogna K, Lifton RP. Boyden LM, et al. N Engl J Med. 2002 May 16;346(20):1513-21. doi: 10.1056/NEJMoa013444. N Engl J Med. 2002. PMID: 12015390 - Pathogenic mutations and polymorphisms in the lipoprotein receptor-related protein 5 reveal a new biological pathway for the control of bone mass.
Ferrari SL, Deutsch S, Antonarakis SE. Ferrari SL, et al. Curr Opin Lipidol. 2005 Apr;16(2):207-14. doi: 10.1097/01.mol.0000162326.62419.e4. Curr Opin Lipidol. 2005. PMID: 15767861 Review. - The genetics of low-density lipoprotein receptor-related protein 5 in bone: a story of extremes.
Balemans W, Van Hul W. Balemans W, et al. Endocrinology. 2007 Jun;148(6):2622-9. doi: 10.1210/en.2006-1352. Epub 2007 Mar 29. Endocrinology. 2007. PMID: 17395706 Review.
Cited by
- Canonical Wnt signaling inhibits osteoclastogenesis independent of osteoprotegerin.
Albers J, Keller J, Baranowsky A, Beil FT, Catala-Lehnen P, Schulze J, Amling M, Schinke T. Albers J, et al. J Cell Biol. 2013 Feb 18;200(4):537-49. doi: 10.1083/jcb.201207142. Epub 2013 Feb 11. J Cell Biol. 2013. PMID: 23401003 Free PMC article. - Update on bone anabolics in osteoporosis treatment: rationale, current status, and perspectives.
Baron R, Hesse E. Baron R, et al. J Clin Endocrinol Metab. 2012 Feb;97(2):311-25. doi: 10.1210/jc.2011-2332. Epub 2012 Jan 11. J Clin Endocrinol Metab. 2012. PMID: 22238383 Free PMC article. Review. - Wnt signaling in bone development and disease: making stronger bone with Wnts.
Regard JB, Zhong Z, Williams BO, Yang Y. Regard JB, et al. Cold Spring Harb Perspect Biol. 2012 Dec 1;4(12):a007997. doi: 10.1101/cshperspect.a007997. Cold Spring Harb Perspect Biol. 2012. PMID: 23209148 Free PMC article. Review. - Modeling craniofacial and skeletal congenital birth defects to advance therapies.
Neben CL, Roberts RR, Dipple KM, Merrill AE, Klein OD. Neben CL, et al. Hum Mol Genet. 2016 Oct 1;25(R2):R86-R93. doi: 10.1093/hmg/ddw171. Epub 2016 Jun 26. Hum Mol Genet. 2016. PMID: 27346519 Free PMC article. Review. - Early chronic kidney disease-mineral bone disorder stimulates vascular calcification.
Fang Y, Ginsberg C, Sugatani T, Monier-Faugere MC, Malluche H, Hruska KA. Fang Y, et al. Kidney Int. 2014 Jan;85(1):142-50. doi: 10.1038/ki.2013.271. Epub 2013 Jul 24. Kidney Int. 2014. PMID: 23884339 Free PMC article.
References
Electronic-Database Information
- Cyrillic program, http://www.cyrillicsoftware.com/ (for visualizing pedigrees)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for human LRP5 cDNA [accession number AF064548], human LRP5 protein [accession number AAC36467], and mouse LRP5 cDNA [accession number AF064984])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for HBM or BMND1 [MIM <601884>], OPS [MIM <259770>], arOP [MIM <259700>]), PYCD [MIM <265800>], DPD1 [MIM 13100], VBCH [MIM <239100>], and melorheostosis [MIM <155950>])
- Protein Data Bank, http://www.rcsb.org/pdb/ (for LDLR YWTD-EGF domain pair [PDB 1IJQ])
- UniGene, http://www.ncbi.nlm.nih.gov/UniGene/ (for UniGene Cluster Hs.116962, CGI-85 [UniGene Cluster Hs.267448], c11orf24 [UniGene Cluster Hs.303025], LRP5 [UniGene Cluster Hs.6347], c11orf23 [UniGene Cluster Hs.180817], MTL5 [UniGene Cluster Hs.121378], GALN [UniGene Cluster Hs.1907], CPT1A [UniGene Cluster Hs.259785], SMBP2 [UniGene Cluster Hs.1521], RTA [UniGene Cluster Hs.118513], [UniGene Cluster Hs.288748], CCND1 [UniGene Cluster Hs.82932], [UniGene Cluster Hs.170932], FGF19 [UniGene Cluster Hs.249200], FGF4 [UniGene Cluster Hs.1755], and FGF3 [UniGene Cluster Hs.37092])
References
- Blank RD (2001) Breaking down bone strength: a perspective on the future of skeletal genetics. J Bone Miner Res 16:1207–1211 - PubMed
- Brown SD, Twells RC, Hey PJ, Cox RD, Levy ER, Soderman AR, Metzker ML, Caskey CT, Todd JA, Hess JF (1998) Isolation and characterization of LRP6, a novel member of the low density lipoprotein receptor gene family. Biochem Biophys Res Commun 248:879–888 - PubMed
- Dausset J, Cann H, Cohen D, Lathrop M, Lalouel JM, White R (1990) Centre d’Etude du Polymorphisme Humain (CEPH): collaborative genetic mapping of the human genome. Genomics 6:575–577 - PubMed
- Del Mastro RG, Lovett M (1997) Isolation of coding sequences from genomic regions using direct selection. Methods Mol Biol 68:183–199 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases