Recombination and mutation during long-term gastric colonization by Helicobacter pylori: estimates of clock rates, recombination size, and minimal age - PubMed (original) (raw)
Recombination and mutation during long-term gastric colonization by Helicobacter pylori: estimates of clock rates, recombination size, and minimal age
D Falush et al. Proc Natl Acad Sci U S A. 2001.
Abstract
The bacterium Helicobacter pylori colonizes the gastric mucosa of half of the human population, resulting in chronic gastritis, ulcers, and cancer. We sequenced ten gene fragments from pairs of strains isolated sequentially at a mean interval of 1.8 years from 26 individuals. Several isolates had acquired small mosaic segments from other H. pylori or point mutations. The maximal mutation rate, the import size, and the frequency of recombination were calculated by using a Bayesian model. The calculations indicate that the last common ancestor of H. pylori existed at least 2,500-11,000 years ago. Imported mosaics have a median size of 417 bp, much smaller than for other bacteria, and recombination occurs frequently (60 imports spanning 25,000 bp per genome per year). Thus, the panmictic population structure of H. pylori results from very frequent recombination during mixed colonization by unrelated strains.
Figures
Figure 1
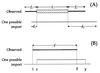
Sequence comparisons of ten gene fragments from 26 pairs of isolates of_H. pylori_. The ten loci are indicated by tiny maps at the top of the figure (colors: core fragments, tan; flanking genes, gray; noncoding regions, white). The sources of the isolates are indicated by patient codes at the left (Colombia, three digits; Louisiana, four digits). The boxes indicate the lengths of the sequenced fragments, distinguished by color (white, identical sequences; yellow, SNPs; pink, clustered polymorphisms). Vertical lines within the boxes indicate the positions of sequence polymorphisms. The patients are separated into five groups (right) on the basis of all genetic changes between the paired isolates (1, no polymorphisms; 2, only SNPs; 3, clustered polymorphisms; 4, related isolates with numerous polymorphic fragments; 5, unrelated isolates). Mathematical analysis was only performed with data from patients in groups 1–3.
Figure 2
Sequence lengths used in the model as described in Materials and Methods.
Figure 3
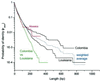
Probability of identity (_p_ident) between sequences versus length. The black curves are pairwise comparisons between the initial isolates within Colombia or Louisiana. The weighted average of these curves is shown in blue. The green curve is from pairwise comparisons between Louisiana and Colombia. Data from within polymorphic stretches from paired isolates (Fig. 1, part 3) are in red.
Figure 4
Parameters estimated by the model for the data in Fig. 1, parts 1–3. (A) Contour plot of marginal likelihoods (posterior/prior) of recombination parameters. (B) Marginal likelihood of mutation rates. The arrow indicates a maximum below which different mutation rates were not distinguished at the 5% level.
Similar articles
- Slow genetic divergence of Helicobacter pylori strains during long-term colonization.
Lundin A, Björkholm B, Kupershmidt I, Unemo M, Nilsson P, Andersson DI, Engstrand L. Lundin A, et al. Infect Immun. 2005 Aug;73(8):4818-22. doi: 10.1128/IAI.73.8.4818-4822.2005. Infect Immun. 2005. PMID: 16040995 Free PMC article. - Of microbe and man: determinants of Helicobacter pylori-related diseases.
van Amsterdam K, van Vliet AH, Kusters JG, van der Ende A. van Amsterdam K, et al. FEMS Microbiol Rev. 2006 Jan;30(1):131-56. doi: 10.1111/j.1574-6976.2005.00006.x. FEMS Microbiol Rev. 2006. PMID: 16438683 Review. - Somatic mutation of mitochondrial DNA in Helicobacter pylori-associated chronic gastritis in patients with and without gastric cancer.
Hiyama T, Tanaka S, Shima H, Kose K, Kitadai Y, Ito M, Sumii M, Yoshihara M, Shimamoto F, Haruma K, Chayama K. Hiyama T, et al. Int J Mol Med. 2003 Aug;12(2):169-74. Int J Mol Med. 2003. PMID: 12851712 - Helicobacter pylori evolution and phenotypic diversification in a changing host.
Suerbaum S, Josenhans C. Suerbaum S, et al. Nat Rev Microbiol. 2007 Jun;5(6):441-52. doi: 10.1038/nrmicro1658. Nat Rev Microbiol. 2007. PMID: 17505524 Review.
Cited by
- In Vivo Genome and Methylome Adaptation of _cag_-Negative Helicobacter pylori during Experimental Human Infection.
Estibariz I, Ailloud F, Woltemate S, Bunk B, Spröer C, Overmann J, Aebischer T, Meyer TF, Josenhans C, Suerbaum S. Estibariz I, et al. mBio. 2020 Aug 25;11(4):e01803-20. doi: 10.1128/mBio.01803-20. mBio. 2020. PMID: 32843556 Free PMC article. - Genome-wide survey of mutual homologous recombination in a highly sexual bacterial species.
Yahara K, Kawai M, Furuta Y, Takahashi N, Handa N, Tsuru T, Oshima K, Yoshida M, Azuma T, Hattori M, Uchiyama I, Kobayashi I. Yahara K, et al. Genome Biol Evol. 2012;4(5):628-40. doi: 10.1093/gbe/evs043. Epub 2012 Apr 25. Genome Biol Evol. 2012. PMID: 22534164 Free PMC article. - Gain and loss of multiple genes during the evolution of Helicobacter pylori.
Gressmann H, Linz B, Ghai R, Pleissner KP, Schlapbach R, Yamaoka Y, Kraft C, Suerbaum S, Meyer TF, Achtman M. Gressmann H, et al. PLoS Genet. 2005 Oct;1(4):e43. doi: 10.1371/journal.pgen.0010043. PLoS Genet. 2005. PMID: 16217547 Free PMC article. - Discovering human history from stomach bacteria.
Disotell TR. Disotell TR. Genome Biol. 2003;4(5):213. doi: 10.1186/gb-2003-4-5-213. Epub 2003 Apr 28. Genome Biol. 2003. PMID: 12734001 Free PMC article. Review. - Mosaic Evolution of Beta-Barrel-Porin-Encoding Genes in Escherichia coli.
Chen X, Cai X, Chen Z, Wu J, Hao G, Luo Q, Liu S, Zhang J, Hu Y, Zhu G, Koester W, White AP, Cai Y, Wang Y. Chen X, et al. Appl Environ Microbiol. 2022 Apr 12;88(7):e0006022. doi: 10.1128/aem.00060-22. Epub 2022 Mar 14. Appl Environ Microbiol. 2022. PMID: 35285711 Free PMC article.
References
- Feldman R A. In: Helicobacter pylori: Molecular and Cellular Biology. Achtman M, Suerbaum S, editors. Norfolk, England: Horizon Scientific Press; 2001. pp. 29–51.
- Achtman M. In: Helicobacter pylori: Molecular and Cellular Biology. Achtman M, Suerbaum S, editors. Wymondham, U.K.: Horizon Scientific Press; 2001. pp. 311–321.
- Achtman M, Azuma T, Berg D E, Ito Y, Morelli G, Pan Z-J, Suerbaum S, Thompson S, van der Ende A, van Doorn L J. Mol Microbiol. 1999;32:459–470. - PubMed
- Kersulyte D, Chalkauskas H, Berg D E. Mol Microbiol. 1999;31:31–43. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases