Methylation at arginine 17 of histone H3 is linked to gene activation - PubMed (original) (raw)
Methylation at arginine 17 of histone H3 is linked to gene activation
Uta-Maria Bauer et al. EMBO Rep. 2002 Jan.
Abstract
The nuclear hormone receptor co-activator CARM1 has the potential to methylate histone H3 at arginine residues in vitro. The methyltransferase activity of CARM1 is necessary for its co-activator functions in transient transfection assays. However, the role of this methyltransferase in vivo is unclear, given that methylation of arginines is not easily detectable on histones. We have raised an antibody that specifically recognizes methylated arginine 17 (R17) of histone H3, the major site of methylation by CARM1. Using this antibody we show that methylated R17 exists in vivo. Chromatin immunoprecipitation analysis shows that R17 methylation on histone H3 is dramatically upregulated when the estrogen receptor-regulated pS2 gene is activated. Coincident with the appearance of methylated R17, CARM1 is found associated with the histones on the pS2 gene. Together these results demonstrate that CARM1 is recruited to an active promoter and that CARM1-mediated R17 methylation on histone H3 takes place in vivo during this active state.
Figures
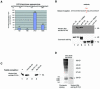
Fig. 1. Methylation of histone H3 at R17 occurs in vivo. (A) Recombinant Drosophila histone H3 was in vitro methylated by GST–CARM1 in the presence of [3H]SAM and subjected to microsequencing of residues 1–30. Amino acid fractions were analysed for the presence of tritium by scintillation counting. Additionally non-radioactively methylated recombinant H3 was sequenced and the amino acid concentration in each fraction was determined by HPLC. The raw data of the radiosequencing were then corrected by calculation of the [3H]methyl incorporation per pmol amino acid. The result of this refinement is shown for the four arginines (R2, R7, R17 and R26) in the H3 N-terminus which are the putative methylation sites of CARM1. The amino acid sequence of residues 1–30 of histone H3 is shown above and the positions of the four arginines are indicated. (B) The antiMe-R17H3 antibody (from
) was raised against a histone H3 peptide (aa 11–24) asymmetrically dimethylated at R17, which represents the major site of CARM1 methylation. Western blot analysis was performed using Me-R17H3 and 2 µg of unmodified recombinant Drosophila histone H3 (lane 1), recombinant H3 after in vitro methylation with GST–CARM1 protein (lane 2), purified calf thymus histone H3 (lane 3), unmodified recombinant Drosophila histone H4 (lane 4), recombinant H4 after in vitro methylation with GST–PRMT1 protein (lane 5) and purified histone H4 (lane 6). Coomassie Blue staining (lower panel) revealed the presence of approximately equal amounts of histones in each lane. The enzymatic activity of GST–CARM1 and GST–PRMT1 on histones was confirmed by in vitro methylation of bulk histones in the presence of [3H-Me]S-adenosyl methionine, SDS–PAGE and autoradiography (data not shown). (C) Peptide competition experiment was performed by western blot analysis using 1 µg of purified calf thymus histone H3 and antiMe-R17H3 antibody in the presence of 1 µg/ml none methylated H3 peptide (aa 11–24, lane 2), R17 methylated H3 peptide (aa 11–24, lane 3) or R3 methylated H4 peptide (aa 1–14, lane 4). (D) Total U2OS cell extract was western blotted using the antiMe-R17H3 antibody. The asterisk indicates methylated histone H3. The left panel shows presence of core histones (indicated on the left) by Coomassie Blue staining. Molecular weights are indicated on the right.
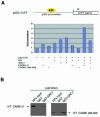
Fig. 2. CARM1 co-activates the pS2 promoter in a methylation dependent manner. (A) 293T cells were transiently transfected with the indicated expression plasmids. Whole-cell extracts were used in CAT assays and the results were quantified on a PhosphorImager. The basal promoter activity of the pS2 CAT reporter in the presence of empty expression vector was normalized to 1.0, and the activities of the remaining transfection reactions were expressed relative to this. The graph shows the result of one experiment which was reproduced independently several times. Equal protein expression of wild-type full-length CARM1 (CARM1 fl) or mutant CARM1 (CARM1 284–608) was confirmed by western blot analysis (data not shown). (B) GST alone or GST fusion protein of SRC1 (984–1441) was expressed in E. coli, purified and used in GST pull-down experiments with in vitro translated (IVT), 35S-labelled CARM1 fl or mutant CARM1 284–608. Interaction between the proteins was analysed by SDS–PAGE and fluorography.
Fig. 3. Activation of the pS2 gene promoter coincides with CARM1 recruitment and methylation of histone H3 at R17 in vivo. MCF-7 cells were stimulated with DMSO/ethanol (no induction control) or with 200 nM 17β-estradiol (E2), or 100 ng/ml tetradecanoyl phorbol acetate (TPA), or both (E2+TPA) for 3 h and analysed by northern blot and ChIP assay. (A) mRNA steady state levels of pS2 and GAPDH were determined by northern blot analysis. Total RNA was prepared from unstimulated and stimulated MCF-7 cells, and 10 µg RNA per sample separated through a 1.2% formaldehyde-agarose gel and blotted onto a nylon membrane. The pS2 and GAPDH RNA was detected by hybridization with 32P-labelled pS2 or GAPDH gene probe. The GAPDH northern blot shows that equal RNA amounts were loaded. (B) For ChIP, genomic chromatin fragments from stimulated MCF-7 cells were immunoprecipitated either with antiMe-R17H3 or antiCARM1 (Upstate) antibodies. Immunoprecipitated chromatin was analysed by quantitative PCR for CARM1 binding and histone H3 R17 methylation in the proximal pS2 promoter region (nt –159 to –463). AntiMe-R17H3 immunoprecipitates were analysed by quantitative PCR with primers for the cdc25 gene promoter (nt –15 to –186). PCR analysis on input chromatin confirmed that equal chromatin amounts were used for ChIPs.
Similar articles
- Histone deimination antagonizes arginine methylation.
Cuthbert GL, Daujat S, Snowden AW, Erdjument-Bromage H, Hagiwara T, Yamada M, Schneider R, Gregory PD, Tempst P, Bannister AJ, Kouzarides T. Cuthbert GL, et al. Cell. 2004 Sep 3;118(5):545-53. doi: 10.1016/j.cell.2004.08.020. Cell. 2004. PMID: 15339660 - Hormone-dependent, CARM1-directed, arginine-specific methylation of histone H3 on a steroid-regulated promoter.
Ma H, Baumann CT, Li H, Strahl BD, Rice R, Jelinek MA, Aswad DW, Allis CD, Hager GL, Stallcup MR. Ma H, et al. Curr Biol. 2001 Dec 11;11(24):1981-5. doi: 10.1016/s0960-9822(01)00600-5. Curr Biol. 2001. PMID: 11747826 - Coactivator-associated arginine methyltransferase-1 enhances nuclear factor-kappaB-mediated gene transcription through methylation of histone H3 at arginine 17.
Miao F, Li S, Chavez V, Lanting L, Natarajan R. Miao F, et al. Mol Endocrinol. 2006 Jul;20(7):1562-73. doi: 10.1210/me.2005-0365. Epub 2006 Feb 23. Mol Endocrinol. 2006. PMID: 16497732 - Bile salt excretory pump: biology and pathobiology.
Suchy FJ, Ananthanarayanan M. Suchy FJ, et al. J Pediatr Gastroenterol Nutr. 2006 Jul;43 Suppl 1:S10-6. doi: 10.1097/01.mpg.0000226385.71859.5f. J Pediatr Gastroenterol Nutr. 2006. PMID: 16819395 Review. - Histone arginine methylation and its dynamic regulation.
Wysocka J, Allis CD, Coonrod S. Wysocka J, et al. Front Biosci. 2006 Jan 1;11:344-55. doi: 10.2741/1802. Front Biosci. 2006. PMID: 16146736 Review.
Cited by
- Phosphoproteomics and lung cancer research.
López E, Cho WC. López E, et al. Int J Mol Sci. 2012 Sep 26;13(10):12287-314. doi: 10.3390/ijms131012287. Int J Mol Sci. 2012. PMID: 23202899 Free PMC article. Review. - ARID2 mitigates hepatic steatosis via promoting the ubiquitination of JAK2.
Cao HJ, Jiang H, Ding K, Qiu XS, Ma N, Zhang FK, Wang YK, Zheng QW, Xia J, Ni QZ, Xu S, Zhu B, Ding XF, Chen TW, Qiu L, Chen W, Li ZG, Zhou B, Feng WM, Xie D, Li JJ. Cao HJ, et al. Cell Death Differ. 2023 Feb;30(2):383-396. doi: 10.1038/s41418-022-01090-0. Epub 2022 Nov 17. Cell Death Differ. 2023. PMID: 36396719 Free PMC article. - Protein arginine methyltransferase 1: positively charged residues in substrate peptides distal to the site of methylation are important for substrate binding and catalysis.
Osborne TC, Obianyo O, Zhang X, Cheng X, Thompson PR. Osborne TC, et al. Biochemistry. 2007 Nov 20;46(46):13370-81. doi: 10.1021/bi701558t. Epub 2007 Oct 26. Biochemistry. 2007. PMID: 17960915 Free PMC article. - Involvement of histone methylation and phosphorylation in regulation of transcription by thyroid hormone receptor.
Li J, Lin Q, Yoon HG, Huang ZQ, Strahl BD, Allis CD, Wong J. Li J, et al. Mol Cell Biol. 2002 Aug;22(16):5688-97. doi: 10.1128/MCB.22.16.5688-5697.2002. Mol Cell Biol. 2002. PMID: 12138181 Free PMC article. - MSK2 and MSK1 mediate the mitogen- and stress-induced phosphorylation of histone H3 and HMG-14.
Soloaga A, Thomson S, Wiggin GR, Rampersaud N, Dyson MH, Hazzalin CA, Mahadevan LC, Arthur JS. Soloaga A, et al. EMBO J. 2003 Jun 2;22(11):2788-97. doi: 10.1093/emboj/cdg273. EMBO J. 2003. PMID: 12773393 Free PMC article.
References
- Aletta J.M., Cimato, T.R. and Ettinger, M.J. (1998) Protein methylation: a signal event in post-translational modification. Trends Biochem. Sci., 23, 89–91. - PubMed
- Bannister A.J., Zegerman, P., Partridge, J.F., Miska, E.A., Thomas, J.O., Allshire, R.C. and Kouzarides, T. (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature, 410, 120–124. - PubMed
- Berger S.L. (2001) An embarrassment of niches: the many covalent modifications of histones in transcriptional regulation. Oncogene, 20, 3007–3013. - PubMed
- Chen D., Ma, H., Hong, H., Koh, S.S., Huang, S.M., Schurter, B.T., Aswad, D.W. and Stallcup, M.R. (1999) Regulation of transcription by a protein methyltransferase. Science, 284, 2174–2177. - PubMed
- Dedon P.C., Soults, J.A., Allis, C.D. and Gorovsky, M.A. (1991) A simplified formaldehyde fixation and immunoprecipitation technique for studying protein–DNA interactions. Anal. Biochem., 197, 83–90. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases