Arp2/3 complex requires hydrolyzable ATP for nucleation of new actin filaments - PubMed (original) (raw)
Arp2/3 complex requires hydrolyzable ATP for nucleation of new actin filaments
M J Dayel et al. Proc Natl Acad Sci U S A. 2001.
Abstract
The Arp2/3 complex, a seven-subunit protein complex containing two actin-related proteins, Arp2 and Arp3, initiates formation of actin filament networks in response to intracellular signals. The molecular mechanism of filament nucleation, however, is not well understood. Arp2 and Arp3 are predicted to bind ATP via a highly conserved nucleotide-binding domain found in all members of the actin superfamily and to form a heterodimer than mimics a conventional actin dimer. We show here that adenosine nucleotides bind with micromolar affinity to both Arp2 and Arp3 and that hydrolyzable ATP is required for actin nucleation activity. Binding of N-WASP WA increases the affinity of both Arp2 and Arp3 for ATP but does not alter the stoichiometry of nucleotides bound in the presence of saturating concentrations of ATP. The Arp2/3 complex bound to ADP or the nonhydrolyzable ATP analogue AMP-PNP cannot nucleate actin filaments, but addition of the phosphate analogue BeF(3) partially restores activity to the ADP-Arp2/3 complex. Bound nucleotide also regulates the affinity of the Arp2/3 complex for its upstream activators N-WASP and ActA. We propose that the active nucleating form of the Arp2/3 complex is the ADP-P(i) intermediate in the ATPase cycle and that the ATPase activity of the Arp2/3 complex controls both nucleation of new filaments and release of the Arp2/3 complex from membrane-associated activators.
Figures
Figure 1
Both the Arp2 and Arp3 subunits of the Arp2/3 complex bind nucleotide with micromolar affinity. (A) UV crosslinking of 5 μM [α-32P]azido-ATP to 1 μM Arp2/3 complex shows both Arp2 and Arp3 bind nucleotide and that [α-32P]azido-ATP crosslinking efficiency is greater on Arp2. (B) Crosslinking of ATP doped with [α-32P]ATP to 2 μM Arp2/3 complex by using UV light. In the presence of 100 μM ATP, radiolabeling occurs mainly on Arp3 with less efficient labeling on Arp2 both in the absence and presence of 25 μM N-WASP WA. (C) Quantitation of_B_ for a range of ATP concentrations fit to the binding quadratic to obtain dissociation constants: Arp3_K_d = 0.6 μM; Arp2_K_d = 1.3 μM; Arp3 + N-WASP WA_K_d = 0.25 μM; Arp2 + N-WASP WA_K_d = 0.5 μM. Binding of N-WASP WA decreases the crosslinking efficiency. (D) Affinity of the Arp2/3 complex for etheno-ATP. Etheno-ATP fluorescence increase upon binding was measured for a range of etheno-ATP concentrations in the presence of a constant concentration of Arp2/3 complex by subtracting the contribution of free etheno-ATP fluorescence from the total measured intensity (see Materials and Methods). Arp2/3 complex alone binds etheno-ATP with an affinity of 3.1 μM, and the N-WASP–Arp2/3 complex binds etheno-ATP with an affinity of 1.3 μM. Binding of N-WASP WA enhances the etheno-ATP fluorescence signal. (E) ATP dissociation kinetics. In 5 μM ATP, nucleotide was driven from 1 μM Arp2/3 complex by mixing with 50 μM etheno-ATP, and binding kinetics were measured by fluorescence enhancement of etheno-ATP on binding. _k_off MgATP = 2.6 s−1. Dissociation slows in the presence of N-WASP WA _k_off MgATP = 0.7 s−1 and enhances the etheno-ATP fluorescence signal.
Figure 2
The affinity of N-WASP WA and ActA for Arp2/3 complex depends on the bound nucleotide. We used polarization anisotropy to measure the affinity of nucleation promoting factors to the Arp2/3 complex in the presence of 1 mM ATP, ADP, and AMP-PNP. Rhodamine ActA (250 nM) (30) binds MgATP-Arp2/3 complex with a_k_d of 3.5 μM, MgADP-Arp2/3 complex with a _k_d of 11.1 μM, and MgAMP-PNP-Arp2/3 complex with a _k_d of 3.0 μM. (Inset) Rhodamine N-WASP WA (250 nM) binds MgATP–Arp2/3 complex with a _k_d of 140 nM and MgADP–Arp2/3 complex with a _k_d of 710 nM.
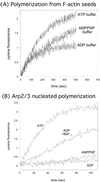
Figure 3
Actin nucleation by Arp2/3 complex requires hydrolyzable ATP. (A) As a control, we tested elongation of 2 μM MgATP-actin from F-actin seeds in the presence of 100 μM ATP, ADP, and AMP-PNP. Kinetics of polymerization are independent of solution nucleotide within the first 100 s of starting the reaction but diverge soon after this, because of the exchange of nucleotide with solution. We therefore limited our observations to the first 100 s when using ATP-actin. (B) The Arp2/3 complex induces polymerization of MgATP-actin in the presence of ATP or ADP-BeF3 but not AMP-PNP or ADP. We used the fact that nucleotide in solution exchanges onto Arp2/3 complex within seconds (Fig. 1_E_) but takes much longer to exchange onto actin monomers (38) to load one type of nucleotide onto the Arp2/3 complex, but keep ATP bound to actin. We mixed 2 μM MgATP-actin with a solution of 10 nM Arp2/3 complex and 1.2 μM N-WASP WA (3-fold above saturating concentration) with 100 μM ATP, ADP, ADP + 2 mM BeF3, or AMP-PNP.
Figure 4
Model for the role of Arp2/3 complex nucleotide binding and hydrolysis in the formation of new actin filaments. Step 1: Assembly of F-actin, Arpd/3 complex, and activator. The Arp2/3 complex binds to the activator, in this case N-WASP, with high affinity because the Arp2/3 complex is ATP-bound. Binding N-WASP brings the actin monomer attached to the WH2 domain of N-WASP in contact with the Arp2/3 complex and this stimulates ATP hydrolysis. Step 2: Hydrolyzing ATP to ADP-Pi causes a conformational change on the complex forming a stable nucleus among Arp3, Arp2, and the conventional actin monomer. Step 3: A new actin filament polymerizes from this nucleus. Step 4: Phosphate release from the Arp2/3 complex decreases its affinity for N-WASP and allows the Arp2/3 complex to release its membrane-associated activator.
Similar articles
- Activation of Arp2/3 complex by Wiskott-Aldrich Syndrome protein is linked to enhanced binding of ATP to Arp2.
Le Clainche C, Didry D, Carlier MF, Pantaloni D. Le Clainche C, et al. J Biol Chem. 2001 Dec 14;276(50):46689-92. doi: 10.1074/jbc.C100476200. Epub 2001 Oct 11. J Biol Chem. 2001. PMID: 11598103 - Activation of Arp2/3 complex: addition of the first subunit of the new filament by a WASP protein triggers rapid ATP hydrolysis on Arp2.
Dayel MJ, Mullins RD. Dayel MJ, et al. PLoS Biol. 2004 Apr;2(4):E91. doi: 10.1371/journal.pbio.0020091. Epub 2004 Apr 13. PLoS Biol. 2004. PMID: 15094799 Free PMC article. - Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex.
Machesky LM, Mullins RD, Higgs HN, Kaiser DA, Blanchoin L, May RC, Hall ME, Pollard TD. Machesky LM, et al. Proc Natl Acad Sci U S A. 1999 Mar 30;96(7):3739-44. doi: 10.1073/pnas.96.7.3739. Proc Natl Acad Sci U S A. 1999. PMID: 10097107 Free PMC article. - Integration of signals to the Arp2/3 complex.
Weaver AM, Young ME, Lee WL, Cooper JA. Weaver AM, et al. Curr Opin Cell Biol. 2003 Feb;15(1):23-30. doi: 10.1016/s0955-0674(02)00015-7. Curr Opin Cell Biol. 2003. PMID: 12517700 Review. - The world according to Arp: regulation of actin nucleation by the Arp2/3 complex.
Welch MD. Welch MD. Trends Cell Biol. 1999 Nov;9(11):423-7. doi: 10.1016/s0962-8924(99)01651-7. Trends Cell Biol. 1999. PMID: 10511705 Review.
Cited by
- Insertions within the actin core of actin-related protein 3 (Arp3) modulate branching nucleation by Arp2/3 complex.
Liu SL, May JR, Helgeson LA, Nolen BJ. Liu SL, et al. J Biol Chem. 2013 Jan 4;288(1):487-97. doi: 10.1074/jbc.M112.406744. Epub 2012 Nov 12. J Biol Chem. 2013. PMID: 23148219 Free PMC article. - Defining coarse-grained representations of large biomolecules and biomolecular complexes from elastic network models.
Zhang Z, Pfaendtner J, Grafmüller A, Voth GA. Zhang Z, et al. Biophys J. 2009 Oct 21;97(8):2327-37. doi: 10.1016/j.bpj.2009.08.007. Biophys J. 2009. PMID: 19843465 Free PMC article. - Force and phosphate release from Arp2/3 complex promote dissociation of actin filament branches.
Pandit NG, Cao W, Bibeau J, Johnson-Chavarria EM, Taylor EW, Pollard TD, De La Cruz EM. Pandit NG, et al. Proc Natl Acad Sci U S A. 2020 Jun 16;117(24):13519-13528. doi: 10.1073/pnas.1911183117. Epub 2020 May 27. Proc Natl Acad Sci U S A. 2020. PMID: 32461373 Free PMC article. - Novel identification of Dermacentor variabilis Arp2/3 complex and its role in rickettsial infection of the arthropod vector.
Petchampai N, Sunyakumthorn P, Guillotte ML, Verhoeve VI, Banajee KH, Kearney MT, Macaluso KR. Petchampai N, et al. PLoS One. 2014 Apr 14;9(4):e93768. doi: 10.1371/journal.pone.0093768. eCollection 2014. PLoS One. 2014. PMID: 24733187 Free PMC article. - Effects of Arp2 and Arp3 nucleotide-binding pocket mutations on Arp2/3 complex function.
Martin AC, Xu XP, Rouiller I, Kaksonen M, Sun Y, Belmont L, Volkmann N, Hanein D, Welch M, Drubin DG. Martin AC, et al. J Cell Biol. 2005 Jan 17;168(2):315-28. doi: 10.1083/jcb.200408177. J Cell Biol. 2005. PMID: 15657399 Free PMC article.
References
- Pollard T, Blanchoin L, Mullins R. Annu Rev Biophys Biomol Struct. 2000;29:545–576. - PubMed
- Taunton J. Curr Opin Cell Biol. 2001;13:85–91. - PubMed
- Ridley A J, Paterson H F, Johnston C L, Diekmann D, Hall A. Cell. 1992;70:401–410. - PubMed
- Ridley A J, Hall A. Cell. 1992;70:389–399. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous