AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation - PubMed (original) (raw)
AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation
Efrat Rabinovich et al. Mol Cell Biol. 2002 Jan.
Abstract
Endoplasmic reticulum-associated degradation (ERAD) disposes of aberrant proteins in the secretory pathway. Protein substrates of ERAD are dislocated via the Sec61p translocon from the endoplasmic reticulum to the cytosol, where they are ubiquitinated and degraded by the proteasome. Since the Sec61p channel is also responsible for import of nascent proteins, this bidirectional passage should be coordinated, probably by molecular chaperones. Here we implicate the cytosolic chaperone AAA-ATPase p97/Cdc48p in ERAD. We show the association of mammalian p97 and its yeast homologue Cdc48p in complexes with two respective ERAD substrates, secretory immunoglobulin M in B lymphocytes and 6myc-Hmg2p in yeast. The membrane 6myc-Hmg2p as well as soluble lumenal CPY*, two short-lived ERAD substrates, are markedly stabilized in conditional cdc48 yeast mutants. The involvement of Cdc48p in dislocation is underscored by the accumulation of ERAD substrates in the endoplasmic reticulum when Cdc48p fails to function, as monitored by activation of the unfolded protein response. We propose that the role of p97/Cdc48p in ERAD, provided by its potential unfoldase activity and multiubiquitin binding capacity, is to act at the cytosolic face of the endoplasmic reticulum and to chaperone dislocation of ERAD substrates and present them to the proteasome.
Figures
FIG. 1.
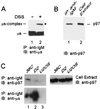
p97/VCP is coprecipitated with unstable μs in B cells. (A) 38C B cells were lysed in the absence (−) (lane 1) or presence (+) (lane 2) of DSS and IgM, and IgM-containing cross-linked complexes were immunoprecipitated with goat anti-IgM antibodies (IP: anti-IgM). Proteins were resolved by SDS-PAGE and were electroblotted onto nitrocellulose, and the blot was probed with rabbit anti-μs followed by HRP-conjugated anti-rabbit IgG (IB: anti-μs). The precipitated μs and ∼100-kDa μs complex are indicated, and the band of ∼100 kDa, marked by the asterisk, was subjected to MALDI-mass spectrometry. (B) 38C B cells were lysed, and IgM and proteins in a complex with IgM were precipitated with goat anti-IgM antibodies. The precipitated proteins (lane 1, IP: anti-IgM) and total cell extract, containing 5% of the proteins subjected to immunoprecipitation (lane 2, cell extract), were resolved by SDS-PAGE and were electroblotted onto nitrocellulose, and the blot was probed with mouse anti-p97 and then with HRP-conjugated anti-mouse IgG (IB: anti-p97). The pulled down p97 is indicated. (C) The μs and associated proteins were precipitated with anti-IgM antibodies (IP: anti-IgM) from 38C (lane 1), D2 (lane 2), and U2OS (lane 3) cells. Proteins were resolved by SDS-PAGE and were electroblotted onto nitrocellulose (left panels). For comparison, total cell extracts (10% of the proteins used for immunoprecipitation) were resolved by SDS-PAGE and were electroblotted onto nitrocellulose (right panel). Upper blots were probed with mouse anti-p97 followed by HRP-conjugated anti-mouse IgG (IB: anti-p97), and the blot in the left panel was reprobed with rabbit anti-μs followed by HRP-conjugated anti-rabbit IgG (IB: anti-μs).
FIG. 2.
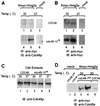
Cdc48p is coprecipitated with 6myc-Hmg2p in yeast. The indicated yeast strains KFY100 (wild-type CDC48) and KFY194 (cdc48-10 ts mutation), transformed with plasmid pRH244 encoding 6myc-Hmg2p (panels A and B, lanes 1 and 2 and lanes 4 and 5; panel C, lanes 1 to 4; panel D, lanes 3 to 5) or mock-transformed (mock) (panels A and B, lanes 3 and 6; panel C, lane 5; panel D, lanes 1 and 2), were grown for 3 h at the indicated temperatures. The 6myc-Hmg2p and associated proteins were precipitated with anti-myc antibodies (IP: anti-myc). Proteins were resolved by SDS-PAGE and were electroblotted onto nitrocellulose. For comparison, total cell extracts (1% of the proteins used for immunoprecipitation) (panel C, lanes 1 to 5) were resolved by SDS-PAGE and were electroblotted onto nitrocellulose. Blots in panels A, C, and D were probed with rabbit anti-Cdc48p followed by HRP-conjugated anti-rabbit IgG (IB: anti-Cdc48p). (B) The blot in panel A was reprobed with mouse anti-myc antibody followed by HRP-conjugated anti-mouse IgG (IB: anti-myc).
FIG. 3.
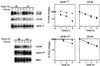
The ERAD of 6myc-Hmg2p is impaired in yeast cdc48 mutants. The stability of 6myc-Hmg2p was measured in strains KFY100 (wild-type CDC48), KFY116 (cdc48-1 cs mutation), and KFY194 (cdc48-10 ts mutation) expressing 6myc-Hmg2p from plasmid pRH244. Strains RHY696 and RHY965 express integrated 6myc-Hmg2p and carry wild-type CDC48, and strain RHY965 is also hrd1Δ. Following preincubation for 2 h at the indicated permissive or restrictive temperatures, cycloheximide (100 μg/ml) was added and cells were collected at the indicated time points. Cells were dissolved and total cellular proteins were resolved by reducing SDS-PAGE and were electroblotted onto nitrocellulose. Blots were probed with mouse anti-myc antibodies followed by HRP-conjugated anti-mouse IgG, and HRP was visualized by ECL. The blots were quantified, and the semi-logarithmic plots represent the decay of 6myc-Hmg2p. The remaining 6myc-Hmg2p was calculated as the percentage of its level at the time of cycloheximide addition (100%). Upper panels, cdc48-1 cs (left) and wild-type CDC48 (right) at 20°C (▴) or 30°C (▪). Lower panels, cdc48-10 ts (left) and wild-type CDC48 (right) at 37°C (•) or 30°C (▪).
FIG. 4.
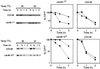
The ERAD of CPY* is impaired in yeast cdc48 mutants. Stability of CPY* was measured for integrated CPY* expressed in strains KFY100 (wild-type CDC48), KFY116 (cdc48-1 cs mutation), and KFY194 (cdc48-10 ts mutation). Following preincubation for 2 h at the indicated permissive or restrictive temperatures, cycloheximide (100 μg/ml) was added and cells were collected at the indicated time points. Cells were dissolved and total cellular proteins were resolved by reducing SDS-PAGE and were electroblotted onto nitrocellulose. Blots were probed with mouse anti-CPY antibodies followed by HRP-conjugated anti-mouse IgG, and the HRP was visualized by ECL. The blots were quantified, and the semi-logarithmic plots represent the decay of CPY*. The remaining CPY* was calculated as the percentage of its level at the time of cycloheximide addition (100%). Upper panels, cdc48-1 cs (left) and wild-type CDC48 (right) at 20°C (▴) or 30°C (▪). Lower panels, cdc48-10 ts (left) and wild-type CDC48 (right) at 37°C (•) or 30°C (▪).
FIG. 5.
The UPR is activated in yeast cdc48 mutants. UPR induction was measured in strains KFY99 and KFY100 carrying wild-type CDC48 or in strain KFY194 carrying the cdc48-10 ts mutation. (A) Activity of β-galactosidase (β-gal) was detected on X-gal reporter plates after a 17-h incubation at the permissive (30°C) or restrictive (37°C) temperatures. (B) β-Galactosidase activity was measured in solubilized cells with ONPG as a substrate after 2.5 to 6 h of incubation at the permissive 30°C (open bars) or restrictive 37°C (lightly shaded bars) temperature. For comparison, cells were incubated at 30°C with tunicamycin (15 μg/ml) (heavily shaded bars). Activity (units) was normalized to the OD600 units of cells used for the assay. The results (means ± standard deviations of 12 independent experiments) are collective data from strains KFY99 and KFY100 (CDC48) or from strain KFY194 (cdc48-10 ts).
Similar articles
- Distinct steps in dislocation of luminal endoplasmic reticulum-associated degradation substrates: roles of endoplamic reticulum-bound p97/Cdc48p and proteasome.
Elkabetz Y, Shapira I, Rabinovich E, Bar-Nun S. Elkabetz Y, et al. J Biol Chem. 2004 Feb 6;279(6):3980-9. doi: 10.1074/jbc.M309938200. Epub 2003 Nov 8. J Biol Chem. 2004. PMID: 14607830 - The role of p97/Cdc48p in endoplasmic reticulum-associated degradation: from the immune system to yeast.
Bar-Nun S. Bar-Nun S. Curr Top Microbiol Immunol. 2005;300:95-125. doi: 10.1007/3-540-28007-3_5. Curr Top Microbiol Immunol. 2005. PMID: 16573238 Review. - Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein.
Huyer G, Piluek WF, Fansler Z, Kreft SG, Hochstrasser M, Brodsky JL, Michaelis S. Huyer G, et al. J Biol Chem. 2004 Sep 10;279(37):38369-78. doi: 10.1074/jbc.M402468200. Epub 2004 Jul 12. J Biol Chem. 2004. PMID: 15252059 - A proteasomal ATPase contributes to dislocation of endoplasmic reticulum-associated degradation (ERAD) substrates.
Lipson C, Alalouf G, Bajorek M, Rabinovich E, Atir-Lande A, Glickman M, Bar-Nun S. Lipson C, et al. J Biol Chem. 2008 Mar 14;283(11):7166-75. doi: 10.1074/jbc.M705893200. Epub 2008 Jan 3. J Biol Chem. 2008. PMID: 18174173 - Endoplasmic reticulum-associated degradation.
Römisch K. Römisch K. Annu Rev Cell Dev Biol. 2005;21:435-56. doi: 10.1146/annurev.cellbio.21.012704.133250. Annu Rev Cell Dev Biol. 2005. PMID: 16212502 Review.
Cited by
- The proteasome cap RPT5/Rpt5p subunit prevents aggregation of unfolded ricin A chain.
Pietroni P, Vasisht N, Cook JP, Roberts DM, Lord JM, Hartmann-Petersen R, Roberts LM, Spooner RA. Pietroni P, et al. Biochem J. 2013 Aug 1;453(3):435-45. doi: 10.1042/BJ20130133. Biochem J. 2013. PMID: 23617410 Free PMC article. - The fine-tuning of proteolytic pathways in Alzheimer's disease.
Cecarini V, Bonfili L, Cuccioloni M, Mozzicafreddo M, Angeletti M, Keller JN, Eleuteri AM. Cecarini V, et al. Cell Mol Life Sci. 2016 Sep;73(18):3433-51. doi: 10.1007/s00018-016-2238-6. Epub 2016 Apr 27. Cell Mol Life Sci. 2016. PMID: 27120560 Free PMC article. Review. - Misfolded proteins traffic from the endoplasmic reticulum (ER) due to ER export signals.
Kincaid MM, Cooper AA. Kincaid MM, et al. Mol Biol Cell. 2007 Feb;18(2):455-63. doi: 10.1091/mbc.e06-08-0696. Epub 2006 Nov 15. Mol Biol Cell. 2007. PMID: 17108324 Free PMC article. - The Arabidopsis SERK1 protein interacts with the AAA-ATPase AtCDC48, the 14-3-3 protein GF14lambda and the PP2C phosphatase KAPP.
Rienties IM, Vink J, Borst JW, Russinova E, de Vries SC. Rienties IM, et al. Planta. 2005 Jun;221(3):394-405. doi: 10.1007/s00425-004-1447-7. Epub 2004 Dec 9. Planta. 2005. PMID: 15592873 - Cytoplasmic localization of the ORF2 protein of hepatitis E virus is dependent on its ability to undergo retrotranslocation from the endoplasmic reticulum.
Surjit M, Jameel S, Lal SK. Surjit M, et al. J Virol. 2007 Apr;81(7):3339-45. doi: 10.1128/JVI.02039-06. Epub 2007 Jan 17. J Virol. 2007. PMID: 17229684 Free PMC article.
References
- Amitay, R., S. Bar-Nun, J. Haimovich, E. Rabinovich, and I. Shachar. 1991. Post-translational regulation of IgM expression in B lymphocytes. J. Biol. Chem. 266:12568–12573. - PubMed
- Amitay, R., I. Shachar, E. Rabinovich, J. Haimovich, and S. Bar-Nun. 1992. Degradation of secretory IgM in B lymphocytes occurs in a post-endoplasmic reticulum compartment and is mediated by a cysteine protease. J. Biol. Chem. 267:20694–20700. - PubMed
- Bays, N. W., R. G. Gardner, L. P. Seelig, C. A. Joazeiro, and R. Y. Hampton. 2001. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat. Cell Biol. 3:24–29. - PubMed
- Biederer, T., C. Volkwein, and T. Sommer. 1997. Role of Cue1p in ubiquitination and degradation at the ER surface. Science 278:1806–1809. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases