T-cell receptor sequences that elicit strong down-regulation of premature termination codon-bearing transcripts - PubMed (original) (raw)
T-cell receptor sequences that elicit strong down-regulation of premature termination codon-bearing transcripts
Jayanthi P Gudikote et al. EMBO J. 2002.
Abstract
The nonsense-mediated decay (NMD) RNA surveillance pathway detects and degrades mRNAs containing premature termination codons (PTCs). T-cell receptor (TCR) and immunoglobulin transcripts, which commonly harbor PTCs as a result of programmed DNA rearrangement during normal development, are down-regulated much more than other known mammalian gene transcripts in response to nonsense codons. Here, we demonstrate that this is not because of promoter or cell type but instead is directed by regulatory sequences within the rearranging VDJ exon and immediately flanking intron sequences of a Vbeta8.1 TCR-beta gene. Insertion of these sequences into a heterologous gene elicited strong down-regulation (>30-fold) in response to PTCs, indicating that this region is sufficient to trigger robust down-regulation. The rearranging Vbeta5.1 exon and the flanking intron sequences from another member of the TCR-beta family also triggered strong down-regulation, suggesting that down-regulatory-promoting elements are a conserved feature of TCR genes. Importantly, we found that the Vbeta8.1 down-regulatory-promoting element was position dependent, such that it failed to function when positioned downstream of a PTC. To our knowledge, this is the first class of down-regulatory elements identified that act upstream of nonsense codons.
Figures
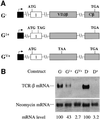
Fig. 1. Cell type and promoter are not responsible for the dramatic down-regulation of TCR-β transcripts in response to nonsense codons. (A) TCR-β and TPI genomic constructs used for transfection. A– and AV+ are PTC– and PTC+ versions, respectively, of the full-length Vβ8.1–Dβ2–Jβ2.3–Cβ2 TCR-β gene, driven by the β-actin promoter. B– and B+ are PTC– and PTC+ versions, respectively, of the TPI gene, driven by the CMV promoter. C– and C+ are PTC– and PTC+ versions, respectively, of the TPI gene, driven by the β-actin promoter. (B) Northern blot analysis of total RNA (10 µg) isolated from NIH 3T3 cells transiently transfected with the constructs shown. The numbers below the blot are relative PTC+ mRNA levels compared with PTC– mRNA levels (PTC– levels are 100) normalized against globin mRNA levels. Comparable results were obtained in two independent transfection experiments.
Fig. 1. Cell type and promoter are not responsible for the dramatic down-regulation of TCR-β transcripts in response to nonsense codons. (A) TCR-β and TPI genomic constructs used for transfection. A– and AV+ are PTC– and PTC+ versions, respectively, of the full-length Vβ8.1–Dβ2–Jβ2.3–Cβ2 TCR-β gene, driven by the β-actin promoter. B– and B+ are PTC– and PTC+ versions, respectively, of the TPI gene, driven by the CMV promoter. C– and C+ are PTC– and PTC+ versions, respectively, of the TPI gene, driven by the β-actin promoter. (B) Northern blot analysis of total RNA (10 µg) isolated from NIH 3T3 cells transiently transfected with the constructs shown. The numbers below the blot are relative PTC+ mRNA levels compared with PTC– mRNA levels (PTC– levels are 100) normalized against globin mRNA levels. Comparable results were obtained in two independent transfection experiments.
Fig. 2. The 5′ portion of TCR-β is responsible for robust down-regulation in response to nonsense codons. (A) Constructs D– and D+ are PTC– and PTC+ versions of the TCR-β mini-gene, respectively. E– and E+ are PTC– and PTC+ versions, respectively, of a TCR-β and TPI chimeric gene comprised of the 5′ half of TCR-β fused to the 3′ half of TPI. F– and F+ are PTC– and PTC+ versions, respectively, of a TCR-β and TPI chimeric gene comprised of the 5′ half of TPI fused to the 3′ half of TCR-β. (B) Northern blot analysis of total RNA (10 µg) isolated from HeLa cells transiently transfected with the constructs shown. The numbers below the blots are relative PTC+ mRNA levels compared with PTC– mRNA levels (PTC– levels are 100). mRNA levels were normalized for differences in transfection efficiency and RNA loading by measurement of the level of neomycin mRNA, which is expressed as a separate transcription unit from all the plasmids in (A). Comparable results were obtained in at least two independent transfection experiments.
Fig. 2. The 5′ portion of TCR-β is responsible for robust down-regulation in response to nonsense codons. (A) Constructs D– and D+ are PTC– and PTC+ versions of the TCR-β mini-gene, respectively. E– and E+ are PTC– and PTC+ versions, respectively, of a TCR-β and TPI chimeric gene comprised of the 5′ half of TCR-β fused to the 3′ half of TPI. F– and F+ are PTC– and PTC+ versions, respectively, of a TCR-β and TPI chimeric gene comprised of the 5′ half of TPI fused to the 3′ half of TCR-β. (B) Northern blot analysis of total RNA (10 µg) isolated from HeLa cells transiently transfected with the constructs shown. The numbers below the blots are relative PTC+ mRNA levels compared with PTC– mRNA levels (PTC– levels are 100). mRNA levels were normalized for differences in transfection efficiency and RNA loading by measurement of the level of neomycin mRNA, which is expressed as a separate transcription unit from all the plasmids in (A). Comparable results were obtained in at least two independent transfection experiments.
Fig. 3. The TCR-β leader exon and its initiation codon are not essential for robust down-regulation in response to nonsense codons. (A) All of the constructs have the Lβ exon from the TCR-β mini-gene replaced with TPI exon 1. G– lacks a PTC, whereas G1+ and GV+ each contain a single nucleotide mutation that generates a PTC in the exons shown. (B) Transfection, northern blot analysis and quantitation were performed as in Figure 2. Comparable results were obtained in two independent transfection experiments.
Fig. 4. The region encompassing a rearranged Vβ8.1DJ exon and flanking intron sequences is essential for strong down-regulation in response to nonsense codons. (A) H– and HC+ are TCR-β constructs that lack the VDJ exon and adjacent intron sequences. H– lacks a PTC, whereas HC+ contains a single nucleotide mutation that creates a PTC in the exon shown. AC+ is a full-length TCR-β construct identical to A– in Figure 1 except that it contains a single nucleotide mutation that creates a PTC in the exon shown. (B) Transfection, northern blot analysis and quantification were performed as in Figure 2. Comparable results were obtained in two independent transfection experiments.
Fig. 4. The region encompassing a rearranged Vβ8.1DJ exon and flanking intron sequences is essential for strong down-regulation in response to nonsense codons. (A) H– and HC+ are TCR-β constructs that lack the VDJ exon and adjacent intron sequences. H– lacks a PTC, whereas HC+ contains a single nucleotide mutation that creates a PTC in the exon shown. AC+ is a full-length TCR-β construct identical to A– in Figure 1 except that it contains a single nucleotide mutation that creates a PTC in the exon shown. (B) Transfection, northern blot analysis and quantification were performed as in Figure 2. Comparable results were obtained in two independent transfection experiments.
Fig. 5. The region containing a rearranged Vβ8.1DJ exon and flanking intron sequences is sufficient to trigger robust down-regulation in response to nonsense codons. (A) All of the constructs have TPI exons 2–4 and flanking intron sequences replaced by the VDJ exon and flanking intron sequences from a rearranged Vβ8.1–Dβ2–Jβ2.3–Cβ2 TCR gene. I– lacks a PTC, whereas IV+ and I6+ each contain a single nucleotide mutation that creates a PTC in the exons shown. I5+ harbors a frameshift mutation in the VDJ exon that creates a PTC in the exon shown. I–ΔJC and IV+ΔJC are the same as I– and IV+, respectively, except that TCR-β IVS-JC intron sequences are deleted. (B) Transfection, northern blot analysis and quantification were performed as in Figure 2. Comparable results were obtained in at least two independent transfection experiments.
Fig. 5. The region containing a rearranged Vβ8.1DJ exon and flanking intron sequences is sufficient to trigger robust down-regulation in response to nonsense codons. (A) All of the constructs have TPI exons 2–4 and flanking intron sequences replaced by the VDJ exon and flanking intron sequences from a rearranged Vβ8.1–Dβ2–Jβ2.3–Cβ2 TCR gene. I– lacks a PTC, whereas IV+ and I6+ each contain a single nucleotide mutation that creates a PTC in the exons shown. I5+ harbors a frameshift mutation in the VDJ exon that creates a PTC in the exon shown. I–ΔJC and IV+ΔJC are the same as I– and IV+, respectively, except that TCR-β IVS-JC intron sequences are deleted. (B) Transfection, northern blot analysis and quantification were performed as in Figure 2. Comparable results were obtained in at least two independent transfection experiments.
Fig. 6. A rearranged Vβ5.1–Jβ1.6 exon and flanking intron sequences promotes strong down-regulation in response to nonsense codons. (A) Both constructs have TPI exons 2–4 and flanking intron sequences replaced with the VJ exon and flanking intron sequences from a rearranged Vβ5.1–Jβ1.6–Cβ1 gene. J– lacks a PTC and J5+ has a frameshift mutation in the VJ exon that creates a PTC in the exon shown. (B) Transfection, northern blot analysis and quantification were performed as in Figure 2. Comparable results were obtained in two independent transfection experiments.
Fig. 7. Position-dependent effect of nonsense codons on the down-regulatory-promoting element. (A) All of the constructs have the Vβ8.1–Dβ2–Jβ2.3 exon and flanking intron sequences moved downstream of the Cβ2.1 exon. K– lacks a PTC, whereas KC+ and KV+ each contain a single nucleotide mutation that creates a PTC in the same positions as in AC+ and AV+, respectively. (B) Transfection, northern blot analysis and quantitation were performed as in Figure 2. Comparable results were obtained in three independent transfection experiments.
Fig. 7. Position-dependent effect of nonsense codons on the down-regulatory-promoting element. (A) All of the constructs have the Vβ8.1–Dβ2–Jβ2.3 exon and flanking intron sequences moved downstream of the Cβ2.1 exon. K– lacks a PTC, whereas KC+ and KV+ each contain a single nucleotide mutation that creates a PTC in the same positions as in AC+ and AV+, respectively. (B) Transfection, northern blot analysis and quantitation were performed as in Figure 2. Comparable results were obtained in three independent transfection experiments.
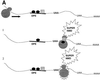
Fig. 8. Models to explain how down-regulatory-promoting elements could stimulate the decay of transcripts containing a downstream nonsense codon. (A) Ribosome sensitization models. Scheme 1, the ribosome picks up positive regulatory proteins from the down-regulatory-promoting element that primes the ribosome for strong NMD when it later encounters a PTC. Scheme 2, the ribosome undergoes a post-translational alteration in response to passage over the down-regulatory-promoting element that primes it for strong NMD when it later encounters a PTC. (B) The cross-talk model, in which the down-regulatory-promoting element communicates with a downstream marker protein to augment its ability to trigger NMD. Abbreviations: DPE, down-regulatory-promoting element; PTSC, post-termination surveillance complex; DSM, downstream mark.
Fig. 8. Models to explain how down-regulatory-promoting elements could stimulate the decay of transcripts containing a downstream nonsense codon. (A) Ribosome sensitization models. Scheme 1, the ribosome picks up positive regulatory proteins from the down-regulatory-promoting element that primes the ribosome for strong NMD when it later encounters a PTC. Scheme 2, the ribosome undergoes a post-translational alteration in response to passage over the down-regulatory-promoting element that primes it for strong NMD when it later encounters a PTC. (B) The cross-talk model, in which the down-regulatory-promoting element communicates with a downstream marker protein to augment its ability to trigger NMD. Abbreviations: DPE, down-regulatory-promoting element; PTSC, post-termination surveillance complex; DSM, downstream mark.
Similar articles
- Efficient downregulation of immunoglobulin mu mRNA with premature translation-termination codons requires the 5'-half of the VDJ exon.
Bühler M, Paillusson A, Mühlemann O. Bühler M, et al. Nucleic Acids Res. 2004 Jun 21;32(11):3304-15. doi: 10.1093/nar/gkh651. Print 2004. Nucleic Acids Res. 2004. PMID: 15210863 Free PMC article. - A splicing-dependent regulatory mechanism that detects translation signals.
Carter MS, Li S, Wilkinson MF. Carter MS, et al. EMBO J. 1996 Nov 1;15(21):5965-75. EMBO J. 1996. PMID: 8918474 Free PMC article. - A quality control pathway that down-regulates aberrant T-cell receptor (TCR) transcripts by a mechanism requiring UPF2 and translation.
Wang J, Vock VM, Li S, Olivas OR, Wilkinson MF. Wang J, et al. J Biol Chem. 2002 May 24;277(21):18489-93. doi: 10.1074/jbc.M111781200. Epub 2002 Mar 11. J Biol Chem. 2002. PMID: 11889124 - [Progress on cis-acting regulatory elements in nonsense-mediated mRNA decay].
Huang Z, Zhou TH, Guo BJ. Huang Z, et al. Yi Chuan Xue Bao. 2004 Nov;31(11):1321-6. Yi Chuan Xue Bao. 2004. PMID: 15651687 Review. Chinese. - Nonsense-mediated mRNA decay modulates clinical outcome of genetic disease.
Khajavi M, Inoue K, Lupski JR. Khajavi M, et al. Eur J Hum Genet. 2006 Oct;14(10):1074-81. doi: 10.1038/sj.ejhg.5201649. Epub 2006 Jun 7. Eur J Hum Genet. 2006. PMID: 16757948 Review.
Cited by
- A 3' UTR sequence stabilizes termination codons in the unspliced RNA of Rous sarcoma virus.
Weil JE, Beemon KL. Weil JE, et al. RNA. 2006 Jan;12(1):102-10. doi: 10.1261/rna.2129806. Epub 2005 Nov 21. RNA. 2006. PMID: 16301601 Free PMC article. - Boundary-independent polar nonsense-mediated decay.
Wang J, Gudikote JP, Olivas OR, Wilkinson MF. Wang J, et al. EMBO Rep. 2002 Mar;3(3):274-9. doi: 10.1093/embo-reports/kvf036. Epub 2002 Feb 15. EMBO Rep. 2002. PMID: 11850396 Free PMC article. - Benchmarking of T cell receptor repertoire profiling methods reveals large systematic biases.
Barennes P, Quiniou V, Shugay M, Egorov ES, Davydov AN, Chudakov DM, Uddin I, Ismail M, Oakes T, Chain B, Eugster A, Kashofer K, Rainer PP, Darko S, Ransier A, Douek DC, Klatzmann D, Mariotti-Ferrandiz E. Barennes P, et al. Nat Biotechnol. 2021 Feb;39(2):236-245. doi: 10.1038/s41587-020-0656-3. Epub 2020 Sep 7. Nat Biotechnol. 2021. PMID: 32895550 - Induced transcription and stability of CELF2 mRNA drives widespread alternative splicing during T-cell signaling.
Mallory MJ, Allon SJ, Qiu J, Gazzara MR, Tapescu I, Martinez NM, Fu XD, Lynch KW. Mallory MJ, et al. Proc Natl Acad Sci U S A. 2015 Apr 28;112(17):E2139-48. doi: 10.1073/pnas.1423695112. Epub 2015 Apr 13. Proc Natl Acad Sci U S A. 2015. PMID: 25870297 Free PMC article. - Frame-disrupting mutations elicit pre-mRNA accumulation independently of frame disruption.
Imam JS, Gudikote JP, Chan WK, Wilkinson MF. Imam JS, et al. Nucleic Acids Res. 2010 Mar;38(5):1559-74. doi: 10.1093/nar/gkp1115. Epub 2009 Dec 9. Nucleic Acids Res. 2010. PMID: 20007599 Free PMC article.
References
- Aoufouchi S., Yélamos,J. and Milstein,C. (1996) Nonsense mutations inhibit RNA splicing in a cell-free system: recognition of mutant codon is independent of protein synthesis. Cell, 85, 415–422. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources