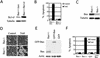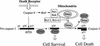TRAIL-induced apoptosis requires Bax-dependent mitochondrial release of Smac/DIABLO - PubMed (original) (raw)
TRAIL-induced apoptosis requires Bax-dependent mitochondrial release of Smac/DIABLO
Yibin Deng et al. Genes Dev. 2002.
Abstract
Recent reports suggest that a cross-talk exists between apoptosis pathways mediated by mitochondria and cell death receptors. In the present study, we report that mitochondrial events are required for apoptosis induced by the cell death ligand TRAIL (TNF-related apoptosis-inducing ligand) in human cancer cells. We show that the Bax null cancer cells are resistant to TRAIL-induced apoptosis. Bax deficiency has no effect on TRAIL-induced caspase-8 activation and subsequent cleavage of Bid; however, it results in an incomplete caspase-3 processing because of inhibition by XIAP. Release of Smac/DIABLO from mitochondria through the TRAIL-caspase-8-tBid-Bax cascade is required to remove the inhibitory effect of XIAP and allow apoptosis to proceed. Inhibition of caspase-9 activity has no effect on TRAIL-induced caspase-3 activation and cell death, whereas expression of the active form of Smac/DIABLO in the cytosol is sufficient to reconstitute TRAIL sensitivity in Bax-deficient cells. Our results show for the first time that Bax-dependent release of Smac/DIABLO, not cytochrome c, from mitochondria mediates the contribution of the mitochondrial pathway to death receptor-mediated apoptosis.
Figures
Figure 1
Requirement of Bax in TRAIL-induced apoptosis. (A) Western analysis of Bcl-xl expression in the Bax+/−/Bcl-xl stable cell line and the control Bax+/−/neo cell line. Whole cell extracts were used. Expression of tubulin was used as a control. (B) Suppression of TRAIL-induced apoptosis in Bax+/−/Bcl-xl cells. Cells were treated with TRAIL for 6 h, apoptosis was measured by trypan blue exclusion, and data from three independent experiments were plotted with standard deviations. (C) Western analysis of Bax expression in Bax+/− and Bax−/− cells. (D) Morphology of TRAIL-treated Bax+/− and Bax−/− cells. The picture was taken 6 h after TRAIL treatment. (E) Comparing the expression of GFP–Bax protein in GFP–Bax stable cell lines derived from Bax−/− cells with Bax expression in Bax+/− cells. Cells expressing GFP only were used as a negative control. Whole cell extracts were analyzed. (F) TRAIL-induced apoptosis in Bax−/−, Bax+/−, GFP, and GFP–Bax cells. Cells were treated with TRAIL (100 ng/mL) for 6 h, apoptosis was measured by trypan blue exclusion, and data from three independent experiments were plotted with standard deviations.
Figure 2
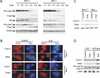
Bax-dependent mitochondrial changes after TRAIL treatment. (A) Activation of caspase-8 and caspase-9 by TRAIL. Whole cell extracts were prepared at different times after TRAIL stimulation as indicated. Western blotting were performed to analyze for cleavage of procaspase-8, procaspase-9, and Bid. Tubulin was used as a loading control. (B) Immunostaining of cytochrome c and Smac/DIABLO. Cells grown on chamber slides were treated by TRAIL for 2 h and stained using anti-cyto c or Smac antibodies after fixing. DNA was visualized by Hoechst 3342. (C) Subcellular fraction of cyto c and Smac/DIABLO. Subcellular fraction was performed on cells before and after TRAIL treatment. The cytosol fraction was subject to Western analysis. Cytosolic β-actin was used as the loading control. (D) Distribution of cyto c and Smac/DIABLO, activation of caspase-9 in Bax reconstituted cells. Cytosol extracts from TRAIL-treated and -untreated GFP- and GFP–Bax-expressing Bax−/− cells were analyzed for cyto c, Smac/DIABLO, and cleavage of caspase-9. Actin is the loading control.
Figure 3
Requirement of Bax translocation in TRAIL-induced apoptosis. (A) Change of Bax distribution after TRAIL treatment. Bax+/− cells were treated by TRAIL, and Bax protein was detected on a Western blot from cytosol and mitochondrial extracts. Cytosol-specific β-actin and mitochondria-specific cytochrome c oxidase subunit IV (coxIV) were used as loading controls. (B) Expression of GFP–BaxΔC21 protein in a GFP–BaxΔC21 stable cell line. Whole cell extracts were used. (C) Bax distribution changes after TRAIL stimulation. (D) Resistance of GFP–BaxΔC21 cells to TRAIL-induced apoptosis. The apoptosis was measured by detection of C-PARP on Western blots.
Figure 4
Protein interaction between caspase-3, XIAP, and Smac/DIABLO, and caspase-3 processing in vivo. (A) Cleavage of procaspase-3 in Bax+/− and Bax−/− cells after TRAIL treatment for 4 h (+) and 24 h. Whole cell extracts were analyzed for procaspase-3, cleaved caspase-3 (p24, p20, p17), and C-PARP on Western blots. (B) Immunoprecipitation of XIAP. The cells were treated by TRAIL for 4 h (indicated by a +) or 2 h. Cytosol extracts were immunoprecipitated with an anti-XIAP antibody and blotted with an anti-caspase-3 antibody. The membrane was stripped and blotted for XIAP later. (C) Smac/DIABLO and XIAP interaction. Cells treated with TRAIL for 4 h and whole cell extracts were examined for Smac/DIABLO and XIAP protein expression on Western blots (WB). Cytosol extracts were immunoprecipitated with anti-XIAP and Smac/DIABLO antibodies, and blotted for Smac/DIABLO and XIAP, respectively.
Figure 5
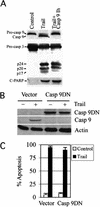
Inhibition of caspase-9 has no effect on caspase-3 processing and TRAIL-mediated apoptosis. (A) Bax+/− cells were treated by TRAIL or a caspase-9 inhibitor (Casp 9Ih) in addition to TRAIL for 4 h. Whole cell extracts were analyzed for caspase-9, caspase-3, and C-PARP by Western blots. (B) Expression of casp 9DN blocks procaspase-9 cleavage. Bax+/− cells were transfected with either the dominant-negative casp 9DN DNA or a vector control along with a GFP-expressing plasmid. More than 90% of the cells were transfected, based on GFP fluorescence. The cells were treated with TRAIL for 4 h at 48 h after transfection. The Casp 9DN protein was detected by an anti-Flag antibody, and processed caspase-9 was detected by an antibody recognizing only the cleaved caspase-9. Actin is used as the loading control. (C) Casp 9DN has no effect on TRAIL-induced apoptosis. Rounded apoptotic GFP-positive cells were counted in triplicate plates, and data were plotted.
Figure 6
Rescue of TRAIL-induced apoptosis. (A) Expression of a full-length Smac/DIABLO cDNA. Bax+/−/Smac and Bax−/−/Smac cells express a full-length Smac/DIABLO (Flag-tagged at the C-terminal) derived from Bax+/− and Bax−/− cells, respectively. They were treated by TRAIL for 4 h. Mitochondrial and cytosol extracts were prepared. Transfected Smac/DIABLO was detected by an anti-Flag antibody, and apoptosis was measured by detection of C-PARP. (B) Expression of the mature active form of Smac/DIABLO in the cytosol of Bax−/− cells. Stable cell lines expressing either GFP or GFP–Smac derived from Bax−/− cells were treated by TRAIL for 4 h, and cytosol extracts were analyzed for expression of GFP, GFP–Smac, and Smac/DIABLO using anti-GFP and Smac/DIABLO antibodies, respectively. Processing and activation of caspase-3 were detected by anti-caspase-3 and C-PARP antibodies. Actin was used as the loading control. (C) Quantitation of apoptosis in GFP and GFP–Smac cells. Apoptosis was measured by trypan blue exclusion; data represent three independent experiments. (D) Detection of Smac/DIABLO–XIAP complex in GFP–Smac-expressing cells. Cytosol extracts as described in B were immunoprecipitated with either anti-XIAP or anti-Smac/DIABLO antibodies, and blotted for Smac/DIABLO or XIAP, respectively.
Figure 7
A model for the role of Smac/DIABLO in cell death receptor-mediated apoptosis.
Similar articles
- Reactive oxygen species regulate caspase activation in tumor necrosis factor-related apoptosis-inducing ligand-resistant human colon carcinoma cell lines.
Izeradjene K, Douglas L, Tillman DM, Delaney AB, Houghton JA. Izeradjene K, et al. Cancer Res. 2005 Aug 15;65(16):7436-45. doi: 10.1158/0008-5472.CAN-04-2628. Cancer Res. 2005. PMID: 16103097 - Regulation of TRAIL-induced apoptosis by ectopic expression of antiapoptotic factors.
Aggarwal BB, Bhardwaj U, Takada Y. Aggarwal BB, et al. Vitam Horm. 2004;67:453-83. doi: 10.1016/S0083-6729(04)67023-3. Vitam Horm. 2004. PMID: 15110190 Review. - Mechanisms of resistance to TRAIL-induced apoptosis in cancer.
Zhang L, Fang B. Zhang L, et al. Cancer Gene Ther. 2005 Mar;12(3):228-37. doi: 10.1038/sj.cgt.7700792. Cancer Gene Ther. 2005. PMID: 15550937 Review.
Cited by
- Fractional killing arises from cell-to-cell variability in overcoming a caspase activity threshold.
Roux J, Hafner M, Bandara S, Sims JJ, Hudson H, Chai D, Sorger PK. Roux J, et al. Mol Syst Biol. 2015 May 7;11(5):803. doi: 10.15252/msb.20145584. Mol Syst Biol. 2015. PMID: 25953765 Free PMC article. - Predicting the cell death responsiveness and sensitization of glioma cells to TRAIL and temozolomide.
Weyhenmeyer BC, Noonan J, Würstle ML, Lincoln FA, Johnston G, Rehm M, Murphy BM. Weyhenmeyer BC, et al. Oncotarget. 2016 Sep 20;7(38):61295-61311. doi: 10.18632/oncotarget.10973. Oncotarget. 2016. PMID: 27494880 Free PMC article. - Impact of p53 status on TRAIL-mediated apoptotic and non-apoptotic signaling in cancer cells.
Willms A, Schittek H, Rahn S, Sosna J, Mert U, Adam D, Trauzold A. Willms A, et al. PLoS One. 2019 Apr 4;14(4):e0214847. doi: 10.1371/journal.pone.0214847. eCollection 2019. PLoS One. 2019. PMID: 30947287 Free PMC article. - Harnessing of Programmed Necrosis for Fighting against Cancers.
Cho YS, Park SY. Cho YS, et al. Biomol Ther (Seoul). 2014 May;22(3):167-75. doi: 10.4062/biomolther.2014.046. Biomol Ther (Seoul). 2014. PMID: 25009696 Free PMC article. Review. - Anticancer activity of sesquiterpenoids extracted from Solanum lyratum via the induction of mitochondria-mediated apoptosis.
Chen M, Wu J, Zhang XX, Wang Q, Yan SH, Wang HD, Liu SL, Zou X. Chen M, et al. Oncol Lett. 2017 Jan;13(1):370-376. doi: 10.3892/ol.2016.5404. Epub 2016 Nov 21. Oncol Lett. 2017. PMID: 28123569 Free PMC article.
References
- Ashkenazi A, Dixit VM. Death receptors: Signaling and modulation. Science. 1998;281:1305–1308. - PubMed
- Bodmer JL, Holler N, Reynard S, Vinciguerra P, Schneider P, Juo P, Blenis J, Tschopp J. TRAIL receptor-2 signals apoptosis through FADD and caspase-8. Nat Cell Biol. 2000;2:241–243. - PubMed
- Burns TF, El-Deiry WS. Identification of inhibitors of TRAIL-induced death (ITIDs) in the TRAIL sensitive colon carcinoma cell line, SW480, using a genetic approach. J Biol Chem. 2001;2:37879–37886. - PubMed
- Cecconi F, Alvarez-Bolado G, Meyer BI, Roth KA, Gruss P. Apaf1 (CED-4 homolog) regulates programmed cell death in mammalian development. Cell. 1998;94:727–737. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials