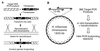A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae - PubMed (original) (raw)
A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae
Brian J Akerley et al. Proc Natl Acad Sci U S A. 2002.
Abstract
A high-density transposon mutagenesis strategy was applied to the Haemophilus influenzae genome to identify genes required for growth or viability. This analysis detected putative essential roles for the products of 259 ORFs of unknown function. Comparisons between complete genomes defined a subset of these proteins in H. influenzae having homologs in Mycobacterium tuberculosis that are absent in Saccharomyces cerevisiae, a distribution pattern that favors their use in development of antimicrobial therapeutics. Three genes within this set are essential for viability in other bacteria. Interfacing the set of essential gene products in H. influenzae with the distribution of homologs in other microorganisms can detect components of unrecognized cellular pathways essential in diverse bacteria. This genome-scale phenotypic analysis identifies potential roles for a large set of genes of unknown function.
Figures
Figure 1
Strategy for whole-genome mutagenesis by in vitro mariner mutagenesis and functional analysis by genetic footprinting. (A) In vitro transposon mutagenesis with Tn_magellan4_ conducted on PCR products corresponding to regions of the H. influenzae chromosome. Each mutagenized DNA segment was introduced separately into H. influenzae by natural transformation and homologous recombination. The Tn_magellan4_ element encodes kanamycin resistance (KmR) and contains a DNA US, both of which are flanked by transposon-inverted repeats (black triangles). (B) PCR products (10 kb) were generated as targets for mutagenesis with ≈5 kb of overlap between adjacent segments. These products were mutagenized (as in A) to generate 366 individual pools of transposon mutants. The same primers were used to obtain genetic footprinting data for every ≈2.5 kb of the genome, and each primer was used on two independent pools of mutants.
Figure 2
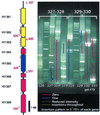
Scoring system for genetic footprinting of the H. influenzae genome. Maps of ORF locations with respect to binding sites of each primer were superimposed on gel images. Dashed (putative nonessential regions) or solid (putative essential regions) lines on the gel image represent predicted segments of H. influenzae coding sequences. Primer reference numbers (red) are listed near primer positions on the gene map (Left) and on the gel image at the bottom of each lane. Tick marks on the gene map represent approximate positions where insertions were detected. Pools of mutants derived from independent transposition reactions are named by the primer pairs used to generate targets for in vitro transposon mutagenesis (top, brackets). The gel image shows analysis of two overlapping ≈10-kb regions corresponding to mutant pools 327–328 and 329–330. Colors denote the summarized results. Red , no insertions; blue, one insertion site; white, insertion mutations partially reduce growth; and yellow, evenly distributed insertions/putative nonessential gene. Absence of insertions within 300 bp of the end of a target PCR product was not considered significant because of the minimum distance required for homologous recombination (e.g., blue line marked with asterisk in lane 327).
Similar articles
- Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung.
Gawronski JD, Wong SM, Giannoukos G, Ward DV, Akerley BJ. Gawronski JD, et al. Proc Natl Acad Sci U S A. 2009 Sep 22;106(38):16422-7. doi: 10.1073/pnas.0906627106. Epub 2009 Sep 4. Proc Natl Acad Sci U S A. 2009. PMID: 19805314 Free PMC article. - Genome scanning in Haemophilus influenzae for identification of essential genes.
Reich KA, Chovan L, Hessler P. Reich KA, et al. J Bacteriol. 1999 Aug;181(16):4961-8. doi: 10.1128/JB.181.16.4961-4968.1999. J Bacteriol. 1999. PMID: 10438768 Free PMC article. - Identification and analysis of essential genes in Haemophilus influenzae.
Wong SM, Akerley BJ. Wong SM, et al. Methods Mol Biol. 2008;416:27-44. doi: 10.1007/978-1-59745-321-9_3. Methods Mol Biol. 2008. PMID: 18392959 Review. - Lack of expression of the global regulator OxyR in Haemophilus influenzae has a profound effect on growth phenotype.
Maciver I, Hansen EJ. Maciver I, et al. Infect Immun. 1996 Nov;64(11):4618-29. doi: 10.1128/iai.64.11.4618-4629.1996. Infect Immun. 1996. PMID: 8890216 Free PMC article. - Genetic systems in Haemophilus influenzae.
Barcak GJ, Chandler MS, Redfield RJ, Tomb JF. Barcak GJ, et al. Methods Enzymol. 1991;204:321-42. doi: 10.1016/0076-6879(91)04016-h. Methods Enzymol. 1991. PMID: 1943781 Review. No abstract available.
Cited by
- Streptococcus pneumoniae TIGR4 Flavodoxin: Structural and Biophysical Characterization of a Novel Drug Target.
Rodríguez-Cárdenas Á, Rojas AL, Conde-Giménez M, Velázquez-Campoy A, Hurtado-Guerrero R, Sancho J. Rodríguez-Cárdenas Á, et al. PLoS One. 2016 Sep 20;11(9):e0161020. doi: 10.1371/journal.pone.0161020. eCollection 2016. PLoS One. 2016. PMID: 27649488 Free PMC article. - Structural basis of the induced-fit mechanism of 1,4-dihydroxy-2-naphthoyl coenzyme A synthase from the crotonase fold superfamily.
Sun Y, Song H, Li J, Li Y, Jiang M, Zhou J, Guo Z. Sun Y, et al. PLoS One. 2013 Apr 26;8(4):e63095. doi: 10.1371/journal.pone.0063095. Print 2013. PLoS One. 2013. PMID: 23658663 Free PMC article. - Essential role of conserved DUF177A protein in plastid 23S rRNA accumulation and plant embryogenesis.
Yang J, Suzuki M, McCarty DR. Yang J, et al. J Exp Bot. 2016 Oct;67(18):5447-5460. doi: 10.1093/jxb/erw311. Epub 2016 Aug 29. J Exp Bot. 2016. PMID: 27574185 Free PMC article. - The ArcA regulon and oxidative stress resistance in Haemophilus influenzae.
Wong SM, Alugupalli KR, Ram S, Akerley BJ. Wong SM, et al. Mol Microbiol. 2007 Jun;64(5):1375-90. doi: 10.1111/j.1365-2958.2007.05747.x. Mol Microbiol. 2007. PMID: 17542927 Free PMC article. - Cloning, purification and preliminary crystallographic analysis of a putative pyridoxal kinase from Bacillus subtilis.
Newman JA, Das SK, Sedelnikova SE, Rice DW. Newman JA, et al. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006 Oct 1;62(Pt 10):1006-9. doi: 10.1107/S1744309106035779. Epub 2006 Sep 30. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006. PMID: 17012797 Free PMC article.
References
- Schena M, Shalon D, Davis R W, Brown P O. Science. 1995;270:467–470. - PubMed
- Mori H, Isono K, Horiuchi T, Miki T. Res Microbiol. 2000;151:121–128. - PubMed
- Ogasawara N. Res Microbiol. 2000;151:129–134. - PubMed
- Smith V, Chou K N, Lashkari D, Botstein D, Brown P O. Science. 1996;274:2069–2074. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 AI026289/AI/NIAID NIH HHS/United States
- AI-48704/AI/NIAID NIH HHS/United States
- R01 AI048704/AI/NIAID NIH HHS/United States
- AI-26289/AI/NIAID NIH HHS/United States
- AI-02137/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources