The Escherichia coli cell division protein FtsW is required to recruit its cognate transpeptidase, FtsI (PBP3), to the division site - PubMed (original) (raw)
The Escherichia coli cell division protein FtsW is required to recruit its cognate transpeptidase, FtsI (PBP3), to the division site
Keri L N Mercer et al. J Bacteriol. 2002 Feb.
Abstract
The bacterial cell division protein FtsW has been suggested to perform two functions: stabilize the FtsZ cytokinetic ring, and facilitate septal peptidoglycan synthesis by the transpeptidase FtsI (penicillin-binding protein 3). We show here that depleting Escherichia coli cells of FtsW had little effect on the abundance of FtsZ rings but abrogated recruitment of FtsI to potential division sites. Analysis of FtsW localization confirmed and extended these results; septal localization of FtsW required FtsZ, FtsA, FtsQ, and FtsL but not FtsI. Thus, FtsW is a late recruit to the division site and is essential for subsequent recruitment of its cognate transpeptidase FtsI but not for stabilization of FtsZ rings. We suggest that a primary function of FtsW homologues--which are found in almost all bacteria and appear to work in conjunction with dedicated transpeptidases involved in division, elongation, or sporulation--is to recruit their cognate transpeptidases to the correct subcellular location.
Figures
FIG. 1.
Recruitment of division proteins to the septal ring of E. coli. The first event is polymerization of FtsZ into the Z-ring. The other proteins localize in the order indicated. For a recent review, see reference . The position of FtsK is from reference . The position of FtsW is from this work.
FIG. 2.
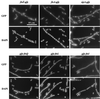
Localization of division proteins in an FtsW depletion background. Strains expressing the indicated GFP fusion were grown in the presence of arabinose or glucose to induce or prevent expression of ftsW, respectively. Cells were fixed and then mixed prior to spreading on slides for microscopy. The short (arabinose-grown) cells often display septal localization of the target division protein and serve as internal controls for microscopy and subsequent image processing. The strains shown are EC856, EC892, EC904, EC860, EC881, and EC861.
FIG. 3.
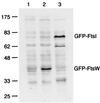
Steady-state levels of GFP-FtsW fusion protein as determined by Western blotting with an anti-GFP antibody. The positions of molecular mass standards (in kilodaltons) and the positions of GFP-FtsW and GFP-FtsI are indicated. Strain EC791 (P209-gfp-ftsW) was grown with 0 mM IPTG (lane 1) or 1 mM IPTG (lane 2). Strain EC436 (P207-gfp-ftsI) was grown with 2.5 μM IPTG (lane 3), which results in production of ≈200 molecules of GFP-FtsI per cell (44). Promoter P209 is much weaker than P207 (44). Chemiluminescence signal was recorded on film, which was then scanned to prepare this figure. In addition, chemiluminescence signal from bands corresponding to the GFP fusion proteins was quantified directly on a Typhoon 8600 imager. The values were 480 for GFP-FtsW uninduced, 3,300 for GFP-FtsW induced, and 3,500 for GFP-FtsI (arbitrary units, corrected for background).
FIG. 4.
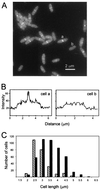
Localization of GFP-FtsW during exponential growth. (A) GFP imaged in a field of cells of strain EC791. (B) Quantitation of the fluorescence in cells that do (a) and do not (b) display septal localization of GFP-FtsW. Transects run lengthwise through the cells indicated in panel A, starting about 1 μm before (lower left) and extending about 1 μm beyond (upper right) each cell. (C) Size distribution of cells that display septal localization of GFP-FtsW. A total of 696 cells were measured and scored for the presence (black bar) or absence (hatched bar) of a fluorescent band extending across the midcell.
FIG. 5.
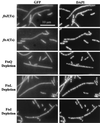
Dependency of GFP-FtsW localization on other Fts proteins. In each case the indicated fts mutant was grown in parallel under permissive and nonpermissive conditions (60 min for temperature-sensitive mutants). Cells were fixed and then mixed prior to spreading on slides for microscopy. The short (permissive condition) cells often display septal localization of GFP-FtsW and serve as internal controls for microscopy and subsequent image processing. The strains shown are EC788, EC787, EC799, EC798, and EC829.
Similar articles
- Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL.
Weiss DS, Chen JC, Ghigo JM, Boyd D, Beckwith J. Weiss DS, et al. J Bacteriol. 1999 Jan;181(2):508-20. doi: 10.1128/JB.181.2.508-520.1999. J Bacteriol. 1999. PMID: 9882665 Free PMC article. - FtsQ, FtsL and FtsI require FtsK, but not FtsN, for co-localization with FtsZ during Escherichia coli cell division.
Chen JC, Beckwith J. Chen JC, et al. Mol Microbiol. 2001 Oct;42(2):395-413. doi: 10.1046/j.1365-2958.2001.02640.x. Mol Microbiol. 2001. PMID: 11703663 - Localization of FtsL to the Escherichia coli septal ring.
Ghigo JM, Weiss DS, Chen JC, Yarrow JC, Beckwith J. Ghigo JM, et al. Mol Microbiol. 1999 Jan;31(2):725-37. doi: 10.1046/j.1365-2958.1999.01213.x. Mol Microbiol. 1999. PMID: 10027987 - Cytokinesis in bacteria.
Errington J, Daniel RA, Scheffers DJ. Errington J, et al. Microbiol Mol Biol Rev. 2003 Mar;67(1):52-65, table of contents. doi: 10.1128/MMBR.67.1.52-65.2003. Microbiol Mol Biol Rev. 2003. PMID: 12626683 Free PMC article. Review. - The structure and function of Escherichia coli penicillin-binding protein 3.
Nguyen-Distèche M, Fraipont C, Buddelmeijer N, Nanninga N. Nguyen-Distèche M, et al. Cell Mol Life Sci. 1998 Apr;54(4):309-16. doi: 10.1007/s000180050157. Cell Mol Life Sci. 1998. PMID: 9614966 Free PMC article. Review.
Cited by
- Structural determinants required to target penicillin-binding protein 3 to the septum of Escherichia coli.
Piette A, Fraipont C, Den Blaauwen T, Aarsman ME, Pastoret S, Nguyen-Distèche M. Piette A, et al. J Bacteriol. 2004 Sep;186(18):6110-7. doi: 10.1128/JB.186.18.6110-6117.2004. J Bacteriol. 2004. PMID: 15342580 Free PMC article. - Regulation of the Peptidoglycan Polymerase Activity of PBP1b by Antagonist Actions of the Core Divisome Proteins FtsBLQ and FtsN.
Boes A, Olatunji S, Breukink E, Terrak M. Boes A, et al. mBio. 2019 Jan 8;10(1):e01912-18. doi: 10.1128/mBio.01912-18. mBio. 2019. PMID: 30622193 Free PMC article. - Two mechanosensitive channel homologs influence division ring placement in Arabidopsis chloroplasts.
Wilson ME, Jensen GS, Haswell ES. Wilson ME, et al. Plant Cell. 2011 Aug;23(8):2939-49. doi: 10.1105/tpc.111.088112. Epub 2011 Aug 2. Plant Cell. 2011. PMID: 21810996 Free PMC article. - Genes required for aerial growth, cell division, and chromosome segregation are targets of WhiA before sporulation in Streptomyces venezuelae.
Bush MJ, Bibb MJ, Chandra G, Findlay KC, Buttner MJ. Bush MJ, et al. mBio. 2013 Sep 24;4(5):e00684-13. doi: 10.1128/mBio.00684-13. mBio. 2013. PMID: 24065632 Free PMC article.
References
- Adam, M., C. Fraipont, N. Rhazi, M. Nguyen-Distèche, B. Lakaye, J. M. Frère, B. Devreese, J. Van Beeumen, Y. van Heijenoort, J. van Heijenoort, and J. M. Ghuysen. 1997. The bimodular G57-V577 polypeptide chain of the class B penicillin-binding protein 3 of Escherichia coli catalyzes peptide bond formation from thiolesters and does not catalyze glycan chain polymerization from the lipid II intermediate. J. Bacteriol. 179:6005-6009. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases