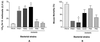Identification of virulence genes in a pathogenic strain of Pseudomonas aeruginosa by representational difference analysis - PubMed (original) (raw)
Identification of virulence genes in a pathogenic strain of Pseudomonas aeruginosa by representational difference analysis
Ji Young Choi et al. J Bacteriol. 2002 Feb.
Abstract
Pseudomonas aeruginosa is an opportunistic pathogen that may cause severe infections in humans and other vertebrates. In addition, a human clinical isolate of P. aeruginosa, strain PA14, also causes disease in a variety of nonvertebrate hosts, including plants, Caenorhabditis elegans, and the greater wax moth, Galleria mellonella. This has led to the development of a multihost pathogenesis system in which plants, nematodes, and insects have been used as adjuncts to animal models for the identification of P. aeruginosa virulence factors. Another approach to identifying virulence genes in bacteria is to take advantage of the natural differences in pathogenicity between isolates of the same species and to use a subtractive hybridization technique to recover relevant genomic differences. The sequenced strain of P. aeruginosa, strain PAO1, has substantial differences in virulence from strain PA14 in several of the multihost models of pathogenicity, and we have utilized the technique of representational difference analysis (RDA) to directly identify genomic differences between P. aeruginosa strains PA14 and PAO1. We have found that the pilC, pilA, and uvrD genes in strain PA14 differ substantially from their counterparts in strain PAO1. In addition, we have recovered a gene homologous to the ybtQ gene from Yersinia, which is specifically present in strain PA14 but absent in strain PAO1. Mutation of the ybtQ homolog in P. aeruginosa strain PA14 significantly attenuates the virulence of this strain in both G. mellonella and a burned mouse model of sepsis to levels comparable to those seen with PAO1. This suggests that the increased virulence of P. aeruginosa strain PA14 compared to PAO1 may relate to specific genomic differences identifiable by RDA.
Figures
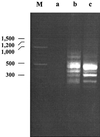
FIG. 1.
Agarose gel electrophoresis (2% gel) of RDA products between P. aeruginosa PA14 and PAO1. The tester DNA amplicons before DNA hybridization and amplification (a) and the difference products after the first (b) and second (c) DNA hybridization-PCR amplification steps are shown. M, Molecular size markers; sizes are on the left, in base pairs.
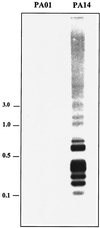
FIG. 2.
Southern blot analysis confirms that the second-round RDA products are unique to PA14. Chromosomal DNA was isolated from P. aeruginosa strains PAO1 and PA14, digested with _Sau_3AI, separated on a 0.8% agarose gel, transferred to a membrane, and hybridized with a labeled pool of the second-round products. Molecular sizes are on the left, in kilobase pairs.
FIG. 3.
Map and nucleotide sequence of ybtQ gene from P. aeruginosa strain PA14. The deduced protein sequence is shown below the coding region of ybtQ. Numbering refers to the letter of the nucleotide sequence. The 196-bp of the RDA product recovered in pJY11 is underlined. The asterisks bracket the ABC transporter domain, and the four boxed regions are conserved motifs of the ABC transporter domain (Walker A, ABC signature, Walker B, and an unnamed fourth motif), as defined by Linton and Higgins (28).
FIG. 4.
Alignment of YbtQ homolog in P. aeruginosa strain PA14 with YbtQ of Y. pestis. An alignment of the YbtQ homolog of P. aeruginosa PA14 with YbtQ of Y. pestis (YP) was generated with Clustal W. Dark boxes indicate identical residues and light boxes enclose similar residues. Numbers above each pair of sequences reflect the deduced protein sequence of the YbtQ homolog in P. aeruginosa PA14.
FIG. 5.
Evaluation of mutants for virulence defects in wax moths and mice. (A) LD50s of P. aeruginosa strains PA14 and PAO1 and four mutants in fifth-instar G. mellonella larvae. Ten larvae were injected at each dilution (containing 0 to 106 bacteria), and larvae were scored as live or dead after 60 h at 25°C. The data are the means and standard deviations of three independent experiments. Asterisks indicate statistically significant differences from P. aeruginosa strain PA14. (B) Mortality in a burned mouse model for P. aeruginosa strains PA14 and PAO1 and for four mutants. Eight mice per experiment were injected subcutaneously with 5 × 105 CFU of each P. aeruginosa strain, and the number of animals that died as a result of sepsis was monitored each day for 10 days. The data are the means and standard deviations of two independent experiments for PA14 and the four mutants; PAO1 was tested once. The asterisk indicates a statistically significant difference from PA14.
Similar articles
- Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial.
Lee DG, Urbach JM, Wu G, Liberati NT, Feinbaum RL, Miyata S, Diggins LT, He J, Saucier M, Déziel E, Friedman L, Li L, Grills G, Montgomery K, Kucherlapati R, Rahme LG, Ausubel FM. Lee DG, et al. Genome Biol. 2006;7(10):R90. doi: 10.1186/gb-2006-7-10-r90. Epub 2006 Oct 12. Genome Biol. 2006. PMID: 17038190 Free PMC article. - Differential roles of the Pseudomonas aeruginosa PA14 rpoN gene in pathogenicity in plants, nematodes, insects, and mice.
Hendrickson EL, Plotnikova J, Mahajan-Miklos S, Rahme LG, Ausubel FM. Hendrickson EL, et al. J Bacteriol. 2001 Dec;183(24):7126-34. doi: 10.1128/JB.183.24.7126-7134.2001. J Bacteriol. 2001. PMID: 11717271 Free PMC article. - Dictyostelium transcriptional responses to Pseudomonas aeruginosa: common and specific effects from PAO1 and PA14 strains.
Carilla-Latorre S, Calvo-Garrido J, Bloomfield G, Skelton J, Kay RR, Ivens A, Martinez JL, Escalante R. Carilla-Latorre S, et al. BMC Microbiol. 2008 Jun 30;8:109. doi: 10.1186/1471-2180-8-109. BMC Microbiol. 2008. PMID: 18590548 Free PMC article. - Elucidating the molecular mechanisms of bacterial virulence using non-mammalian hosts.
Mahajan-Miklos S, Rahme LG, Ausubel FM. Mahajan-Miklos S, et al. Mol Microbiol. 2000 Sep;37(5):981-8. doi: 10.1046/j.1365-2958.2000.02056.x. Mol Microbiol. 2000. PMID: 10972817 Review. - Pseudomonas aeruginosa reference strains PAO1 and PA14: A genomic, phenotypic, and therapeutic review.
Grace A, Sahu R, Owen DR, Dennis VA. Grace A, et al. Front Microbiol. 2022 Oct 13;13:1023523. doi: 10.3389/fmicb.2022.1023523. eCollection 2022. Front Microbiol. 2022. PMID: 36312971 Free PMC article. Review.
Cited by
- Caenorhabditis elegans as a model host for Staphylococcus aureus pathogenesis.
Sifri CD, Begun J, Ausubel FM, Calderwood SB. Sifri CD, et al. Infect Immun. 2003 Apr;71(4):2208-17. doi: 10.1128/IAI.71.4.2208-2217.2003. Infect Immun. 2003. PMID: 12654843 Free PMC article. - Bacteriophages and phage-derived proteins--application approaches.
Drulis-Kawa Z, Majkowska-Skrobek G, Maciejewska B. Drulis-Kawa Z, et al. Curr Med Chem. 2015;22(14):1757-73. doi: 10.2174/0929867322666150209152851. Curr Med Chem. 2015. PMID: 25666799 Free PMC article. Review. - The broad host range pathogen Pseudomonas aeruginosa strain PA14 carries two pathogenicity islands harboring plant and animal virulence genes.
He J, Baldini RL, Déziel E, Saucier M, Zhang Q, Liberati NT, Lee D, Urbach J, Goodman HM, Rahme LG. He J, et al. Proc Natl Acad Sci U S A. 2004 Feb 24;101(8):2530-5. doi: 10.1073/pnas.0304622101. Proc Natl Acad Sci U S A. 2004. PMID: 14983043 Free PMC article. - The Yersiniabactin-Associated ATP Binding Cassette Proteins YbtP and YbtQ Enhance Escherichia coli Fitness during High-Titer Cystitis.
Koh EI, Hung CS, Henderson JP. Koh EI, et al. Infect Immun. 2016 Apr 22;84(5):1312-1319. doi: 10.1128/IAI.01211-15. Print 2016 May. Infect Immun. 2016. PMID: 26883590 Free PMC article. - Genomic Differences Associated with Resistance and Virulence in Pseudomonas aeruginosa Isolates from Clinical and Environmental Sites.
Aroca Molina KJ, Gutiérrez SJ, Benítez-Campo N, Correa A. Aroca Molina KJ, et al. Microorganisms. 2024 May 30;12(6):1116. doi: 10.3390/microorganisms12061116. Microorganisms. 2024. PMID: 38930498 Free PMC article.
References
- Ames, G. F., C. S. Mimura, S. R. Holbrook, and V. Shyamala. 1992. Traffic ATPases: a superfamily of transport proteins operating from Escherichia coli to humans. Adv. Enzymol. Relat. Areas Mol. Biol. 65:1-47. - PubMed
- Baltch, A. L. 1994. Pseudomonas bacteremia, p. 73-128. In R. P. Smith and A. L. Baltch (ed.), Pseudomonas aeruginosa infections and treatment. Marcel Dekker, New York, N.Y.
- Bayer, A. S., and D. C. Norman. 1990. Valve site-specific pathogenetic differences between right-sided and left-sided bacterial endocarditis. Chest 98:200-205. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources