Localization of large ADP-ribosylation factor-guanine nucleotide exchange factors to different Golgi compartments: evidence for distinct functions in protein traffic - PubMed (original) (raw)
Comparative Study
Localization of large ADP-ribosylation factor-guanine nucleotide exchange factors to different Golgi compartments: evidence for distinct functions in protein traffic
Xinhua Zhao et al. Mol Biol Cell. 2002 Jan.
Abstract
Activation of several ADP-ribosylation factors (ARFs) by guanine nucleotide exchange factors (GEFs) regulates recruitment of coat proteins (COPs) on the Golgi complex and is generally assumed to be the target of brefeldin A (BFA). The large ARF-GEFs Golgi-specific BFA resistance factor 1 (GBF1) and BFA-inhibited GEFs (BIGs) localize to this organelle but catalyze exchange preferentially on class II and class I ARFs, respectively. We now demonstrate using quantitative confocal microscopy that these GEFs show a very limited overlap with each other (15 and 23%). In contrast, GBF1 colocalizes with the cis-marker p115 (86%), whereas BIGs overlap extensively with TGN38 (83%). Consistent with these distributions, GBF1, but not BIG1, partially relocalized to peripheral sites after incubation at 15 degrees C. The new GBF1 structures represent peripheral vesicular tubular clusters (VTCs) because 88% of structures analyzed stained for both GBF1 and p115. Furthermore, as expected of VTCs, they rapidly reclustered to the Golgi complex in a microtubule-dependent manner upon warm-up. These observations suggest that GBF1 and BIGs activate distinct subclasses of ARFs in specific locations to regulate different types of reactions. In agreement with this possibility, COPI overlapped to a greater extent with GBF1 (64%) than BIG1 (31%), whereas clathrin showed limited overlap with BIG1, and virtually none with GBF1.
Figures
Figure 1
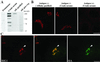
Endogenous BIG1 localizes to the Golgi complex through sequences present in the N-terminal third of the protein. (A) Both crude and affinity-purified 9D3 serum (anti-BIG1) label a protein of 208 kDa, the size predicted from the cDNA of hBIG1. NRK cell lysates (400 μg) was loaded into the wide lane (7.3 cm) of an 8% gel, separated by SDS-PAGE, and then transferred to a nitrocellulose membrane. Membrane strips (0.5 cm in width) were incubated with the indicated sera at equivalent dilutions and processed for enhanced chemiluminescence. One strip was incubated with antibody in the presence of 2 μg antigen, as indicated. The position of molecular weight standards run in parallel is shown on the right. Shown are representative data from two experiments with similar results. (B) Both crude and affinity-purified 9D3 serum specifically localize endogenous BIG1 to a perinuclear structure by IF. NRK cells were fixed and processed for IF by incubation with the indicated 9D3 sera (crude serum or purified antibody) in the absence or presence of the immunizing antigen (1 μg). Shown are single slice confocal images. (C) N-Terminal third fragment of BIG1 (N1- BIG1) is recruited to the same subcompartment within the Golgi complex as full-length endogenous BIG1. NRK cells were transiently transfected with constructs encoding HA- tagged N1 fragment of BIG1 and processed for IF by incubation with rabbit serum 9D3 and monoclonal anti-HA 3F10. Cell that has been transfected is indicated by arrowhead. Images were obtained by standard epifluorescence microscopy because the limited number of transfectants made confocal microscopy impractical in this case. The middle image was taken using a dichroic filter to simultaneously capture the signal from both the red and green fluorophores. Bars (B and C), 10 μm.
Figure 2
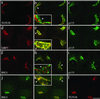
GBF1 and BIG1 are recruited to different compartments of the Golgi complex. (a) BHK-21 cells were transiently transfected with a plasmid encoding HA-tagged full-length BIG1 and processed for IF by using antibodies against the HA epitope (3F10/fluorescein) and GBF1 (H154/Texas Red). (b) NRK cells were fixed and processed for IF by using antibodies against GBF1 (H154/Alexa594) and BIG1 (Alexa488-conjugated 9D3). Both images (a and b) were obtained by collecting stacks of optical sections (3.2 μm in thickness for BHK-21 cell [a] and 2.0 μm for NRK cell [b]) by using a confocal microscope. Shown in the center is an X-Y slice. Above and to the right are slices through the stack that reveal the distribution in the X-Z and Y-Z planes at the position indicated by the magenta and green lines, respectively. Bars, 10 μm. Similar results were obtained from at least three independent experiments.
Figure 3
GBF1 and BIG1 localize to_cis_- and _trans_-compartments of the Golgi complex, respectively. NRK cells were fixed and processed for double-label IF by using either polyclonal anti-TGN38 (a), anti-GBF1 (d), or anti-BIG1 (g), and monoclonal anti-p115 (c, f, and i), or the two polyclonal antibodies Alexa488-conjugated 9D3 (anti-BIG1, j) and anti-TGN38 (l). Shown are single slice confocal images. Middle panels (b, e, h, and k) show merged left and right images. The inset shows a threefold magnification of the area indicated by an arrowhead. Bar, 10 μm. Shown are representative data from at least three experiments with similar results.
Figure 4
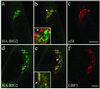
N terminal fragment of BIG2, like BIG1, does not colocalize with GBF1 and the_cis_-Golgi marker p58. BHK-21 cells were transiently transfected with a plasmid encoding HA-tagged N- BIG2 and processed for double-label IF by using monoclonal anti-HA and polyclonal antibodies against either p58 (a–c) or GBF1 (d–f). Shown are single slice confocal images obtained with the indicated antibody. Middle panels (b and e) show superimposed left and right images. The inset shows a threefold magnification of the area indicated by an arrowhead. Bars, 5 μm. Distinct staining for HA-BIG2 and GBF1 was observed in each of four experiments.
Figure 5
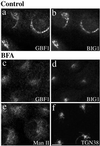
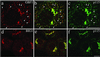
GBF1 and BIG1 redistribute to distinct structures in response to BFA. NRK cells were treated with either 0.1% DMSO (control) or 10 μg/ml BFA for 30 min at 37°C before fixation and processing for IF. Cells were either double stained using two polyclonal antibodies (a–d) against GBF1 (H154/Alexa594; a and c) and BIG1 (Alexa488-conjugated 9D3; b and d) or singly stained using polyclonal antibodies against Man II (e) or TGN38 (f). Shown are single-slice confocal images. Bar, 10 μm. Similar results were obtained from at least two independent experiments.
Figure 6
GBF1, but not BIG1, redistributes from the Golgi complex to peripheral VTCs at 15°C. NRK cells incubated for 2 h in a 15°C water bath before fixation were processed for double-label IF by using monoclonal antibodies against p115 and polyclonal antibodies that recognize either GBF1 (a–c) or BIG1 (d–f). To better reveal small peripheral structures, projections of several confocal slices are shown. Middle panels (b and e) show superimposed left and right images. As indicated by arrowheads in images a–c, the peripheral GBF1 and p115 staining patterns are almost identical. Peripheral structures appear as either yellow or orange dots depending on the relative intensity of red and green signals. The apparent partial overlap between BIG1 and p115 in perinuclear area shown in image e is due to the overexposure of perinuclear signals to better display small peripheral structures. Bars, 10 μm. The distinct response to temperature shift for GBF1 and BIG1 was observed in each of at least three separate experiments.
Figure 7
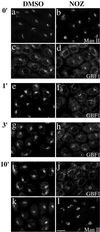
Redistribution of GBF1 from the 15°C peripheral compartments upon warm-up to 37°C is microtubule dependent. NRK cells were transferred to DMEM containing HEPES pH 7.4 and either 0.1% DMSO vehicle control or 5 μg/ml nocodazole (NOZ) and then incubated in a 15°C water bath for 2 h. Cells were either immediately fixed (0′) or quickly transferred to 37°C water bath and incubated for additional 1 min (1′), 3 min (3′), or 10 min (10′) before fixation. Coverslips were processed for IF by using polyclonal anti-GBF1 (e–h) or double-label IF (a–d and i–l) by using polyclonal anti- GBF1 (c and d, and I and j) and monoclonal anti-Man II (53FC3) (a and b, and k and l). Images obtained by standard epifluorescence microscopy are presented. Bar, 10 μm. Similar results were obtained in at least two independent experiments.
Figure 8
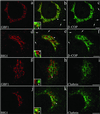
GBF1 and BIG1 overlap with distinct sets of coat proteins: GBF1 significantly with β-COP and BIG1 preferentially with clathrin. NRK cells were fixed and processed for double-label IF by using anti-β-COP and anti-GBF1 (a–c), anti-β-COP and anti-BIG1 (d–f), anti- clathrin and anti-GBF1 (g–i), or anti-clathrin and anti- BIG1 (j–l). Shown are single-slice confocal images taken in the indicated channel. Middle panels (b, e, h, and k) show superimposed left and right images. The inset shows a fourfold magnification of the area indicated by arrowhead. Arrows in b, c, e, and f point to some peripheral structures stained by anti-β-COP only. Bars, 5 μm. Shown are representative data from at least two experiments.
Figure 9
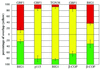
Quantitative analysis of the distribution of ARF-GEFs relative to Golgi markers and coat proteins. The extent of overlap observed in the perinuclear area by using antibodies to the indicated proteins was measured as described in MATERIALS AND METHODS. The percentage of IF signal intensity present in shared pixels (yellow) relative to total signal was measured for both the red (top) and green (bottom) channels. Red and green only values were defined as the difference between 100% and the overlap measured for the corresponding channel. Red signal was obtained by staining with polyclonal anti-GBF1 or anti- BIG1, whereas green signal was obtained with monoclonal anti-p115, anti-β-COP, or Alexa488-conjugated polyclonal anti-BIG1 (see legends of Figures 2, 3, and 8 for details). The number of separate images used for quantitation was five for GBF1/p115, four for TGN38/BIG1, five for GBF1/BIG1, six for GBF1/β-COP, and six for BIG1/β-COP. Error bars correspond to the SD of the mean.
Similar articles
- GBF1, a cis-Golgi and VTCs-localized ARF-GEF, is implicated in ER-to-Golgi protein traffic.
Zhao X, Claude A, Chun J, Shields DJ, Presley JF, Melançon P. Zhao X, et al. J Cell Sci. 2006 Sep 15;119(Pt 18):3743-53. doi: 10.1242/jcs.03173. Epub 2006 Aug 22. J Cell Sci. 2006. PMID: 16926190 - GBF1, a guanine nucleotide exchange factor for ADP-ribosylation factors, is localized to the cis-Golgi and involved in membrane association of the COPI coat.
Kawamoto K, Yoshida Y, Tamaki H, Torii S, Shinotsuka C, Yamashina S, Nakayama K. Kawamoto K, et al. Traffic. 2002 Jul;3(7):483-95. doi: 10.1034/j.1600-0854.2002.30705.x. Traffic. 2002. PMID: 12047556 - GBF1: A novel Golgi-associated BFA-resistant guanine nucleotide exchange factor that displays specificity for ADP-ribosylation factor 5.
Claude A, Zhao BP, Kuziemsky CE, Dahan S, Berger SJ, Yan JP, Armold AD, Sullivan EM, Melançon P. Claude A, et al. J Cell Biol. 1999 Jul 12;146(1):71-84. J Cell Biol. 1999. PMID: 10402461 Free PMC article. - GBF1 and Arf1 function in vesicular trafficking, lipid homoeostasis and organelle dynamics.
Kaczmarek B, Verbavatz JM, Jackson CL. Kaczmarek B, et al. Biol Cell. 2017 Dec;109(12):391-399. doi: 10.1111/boc.201700042. Epub 2017 Nov 6. Biol Cell. 2017. PMID: 28985001 Review. - Activation of toxin ADP-ribosyltransferases by eukaryotic ADP-ribosylation factors.
Moss J, Vaughan M. Moss J, et al. Mol Cell Biochem. 1999 Mar;193(1-2):153-7. Mol Cell Biochem. 1999. PMID: 10331652 Review.
Cited by
- Toward a model for Arf GTPases as regulators of traffic at the Golgi.
Kahn RA. Kahn RA. FEBS Lett. 2009 Dec 3;583(23):3872-9. doi: 10.1016/j.febslet.2009.10.066. Epub 2009 Oct 29. FEBS Lett. 2009. PMID: 19879269 Free PMC article. Review. - The GTPase Arf1p and the ER to Golgi cargo receptor Erv14p cooperate to recruit the golgin Rud3p to the cis-Golgi.
Gillingham AK, Tong AH, Boone C, Munro S. Gillingham AK, et al. J Cell Biol. 2004 Oct 25;167(2):281-92. doi: 10.1083/jcb.200407088. J Cell Biol. 2004. PMID: 15504911 Free PMC article. - Rab1b interacts with GBF1 and modulates both ARF1 dynamics and COPI association.
Monetta P, Slavin I, Romero N, Alvarez C. Monetta P, et al. Mol Biol Cell. 2007 Jul;18(7):2400-10. doi: 10.1091/mbc.e06-11-1005. Epub 2007 Apr 11. Mol Biol Cell. 2007. PMID: 17429068 Free PMC article. - A C-terminal sequence in the guanine nucleotide exchange factor Sec7 mediates Golgi association and interaction with the Rsp5 ubiquitin ligase.
Dehring DA, Adler AS, Hosseini A, Hicke L. Dehring DA, et al. J Biol Chem. 2008 Dec 5;283(49):34188-96. doi: 10.1074/jbc.M806023200. Epub 2008 Oct 2. J Biol Chem. 2008. PMID: 18832381 Free PMC article. - Distinct functions for Arf guanine nucleotide exchange factors at the Golgi complex: GBF1 and BIGs are required for assembly and maintenance of the Golgi stack and trans-Golgi network, respectively.
Manolea F, Claude A, Chun J, Rosas J, Melançon P. Manolea F, et al. Mol Biol Cell. 2008 Feb;19(2):523-35. doi: 10.1091/mbc.e07-04-0394. Epub 2007 Nov 14. Mol Biol Cell. 2008. PMID: 18003980 Free PMC article.
References
- Antonny B, Schekman R. ER export: public transportation by the COPII coach. Curr Opin Cell Biol. 2001;13:438–443. - PubMed
- Brown HA, Gutowsk S, Moomaw CR, Slaughter C, Sternweis PC. ADP- ribosylation factor, a small GTP-dependent regulatory protein, stimulates phospholipase D activity. Cell. 1993;75:1137–1144. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases