Transcription factor Sp3 is regulated by acetylation - PubMed (original) (raw)
Transcription factor Sp3 is regulated by acetylation
H Braun et al. Nucleic Acids Res. 2001.
Abstract
Sp3 is a ubiquitous transcription factor closely related to Sp1. Previous analyses showed that, unlike Sp1, Sp3 fails to activate transcription in certain promoter settings. This is due to the presence of an inhibitory domain located between the second glutamine-rich activation domain and the DNA-binding domain. To further analyze the transcriptional properties of Sp3, we have expressed and purified recombinant Sp3 and Sp1 as epitope-tagged proteins from stable transfected insect cells. We found that Sp3 does act as a strong activator similar to Sp1 in an in vitro transcription assay using Sp1/Sp3-depleted HeLa nuclear extract. However, on the same promoter Sp3 is almost inactive when transfected into cells. Mutational studies demonstrate that a single lysine residue is responsible for the low transcriptional activity of Sp3 in vivo. We show that Sp3, but not a mutant of Sp3 that lacks this lysine residue, is highly acetylated in vivo. Our results strongly suggest that the transcriptional activity of Sp3 is regulated by acetylation. The consequences of acetylation for the activity of Sp3 are discussed.
Figures
Figure 1
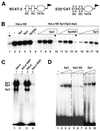
Expression and purification of epitope-tagged Sp3 and Sp3SD in stable transfected SL2 cells. (A) Schematic representation of Sp1, Sp3 and the Sp3 mutant Sp3SD expressed in SL2 cells. Gray boxes indicate the glutamine-rich activation domains, three black bars the three zinc fingers of the Sp factors. The box with hatched stripes N-terminal to the zinc fingers in Sp3 depicts the inhibitory domain. All three proteins contain a hemagglutinin (HA) and a FLAG epitope at their N-termini. (B) Induction of epitope-tagged Sp3 in stable transfected SL2 cells. Expression of Sp3 was induced with 500 µM copper sulfate. At various times after induction (0.5–36 h) nuclear extracts were prepared and subjected to electrophoretic mobility shift analysis. In lanes 1–7 0.75 µg and in lanes 8–12 1.2 µg extracted protein were used for the GC box-binding reaction. (C) Detection of epitope-tagged Sp3 by western blotting. Extracts of epitope-tagged Sp3-expressing SL2 cells were subjected to western blot analyses using antibodies against the FLAG epitope (αFLAG), the HA epitope (αHA) and two different anti-Sp3 antibodies [αSp3 (3) and αSp3c (Santa Cruz)]. (D) Purification of epitope-tagged Sp3 from stable transfected SL2 cells using immune affinity chromatography. Aliquots of the various purification steps were analyzed by SDS–PAGE and stained with Coomassie blue. M, marker lane; NE, crude nuclear extract; UN, breakthrough of a non-specific immune affinity chromatography (preimmune αSp3 serum coupled to protein A–Sepharose); BT, breakthrough of a specific immune affinity chromatography (αFLAG matrix); Elution, consecutive 300 µl fractions upon elution with FLAG peptide.
Figure 2
In vitro transcription with purified recombinant Sp1, Sp3 and Sp3SD. (A) Schematic representation of the reporter plasmids BCAT-2 and E(II)*CAT used as templates. BCAT-2 contains two adjacent GC boxes fused to the E1B TATA-box. Both GC boxes were mutated in E(II)*CAT. (B) Primer extension analyses of in vitro transcribed RNA. In vitro transcription reactions were performed with HeLa nuclear extract programmed with BCAT-2 or E(II)*CAT (the latter being indicated by black circles, lanes 1, 10 and 14) as templates. Increasing amounts of purified recombinant Sp3 (2, 4 and 8 µl, lanes 7–10), Sp3SD (2, 4 and 8 µl, lanes 11–14) or Sp1 (1, 2, 4 and 8 µl, lanes 15–18) were added to whole HeLa nuclear extract (lanes 1–5) or HeLa nuclear extract depleted of GC box-binding proteins (lanes 6–18). Reaction products were subjected to denaturing PAGE and autoradiography. (C) GC box-binding activity in HeLa nuclear extracts used for in vitro transcription assays. HeLa nuclear extract (1 and 3 µl) prior to (lanes 1 and 3) and after (lanes 2 and 4) incubation with a GC box affinity matrix was incubated with 0.1 ng 32P-labeled GC box oligonucleotide and subjected to EMSA. (D) Purified recombinant Sp3, Sp3SD and Sp1 fractions used for in vitro transcription experiments were analyzed by EMSA. Binding reactions contained 0.05 µl (lanes 2, 5 and 8), 0.075 µl (lanes 3, 6 and 9) or 0.1 µl (lanes 4, 7 and 10) of the protein extracts used for the transcription reactions shown in (B).
Figure 3
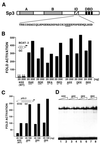
Point mutations in the inhibitory domain strongly enhance Sp3 activation capacity. (A) Schematic drawing of Sp3. A and B indicate the glutamine-rich activation domains, ID the inhibitory domain and DBD the zinc finger DNA-binding domain. Double and single point mutations were introduced into the intact Sp3 molecule within the KEE sequence of the inhibitory domain. (B) SL2 cells were transfected with 4 µg BCAT-2 plasmid along with 20 and 200 ng expression plasmids for wild-type Sp3 or Sp3 mutants, respectively, as indicated. Amino acids that differ from the wild-type KEE sequence are underlined. The CAT values are expressed relative to the CAT activity obtained with the vector (pPac), which has been given the arbitrary value of 1. The values represent mean values of at least two independent transfections. (C) SL2 cells were transfected with 20 and 100 ng expression plasmids for the Sp3 lysine mutants along with 4 µg SV40 promoter-driven luciferase reporter plasmid pGL3. (D) Transient expression of Sp3 mutant proteins in SL2 cells. Gel retardation assays were performed with crude nuclear extracts from SL2 cells transfected with 10 µg expression plasmids for wild-type Sp3 (KEE, lanes 1 and 2) or the lysine mutants (
A
EE, lanes 3 and 4;
D
EE, lanes 5 and 6;
R
EE, lanes 7 and 8) as indicated. All reactions contained 0.2 ng 32P-labeled GC box oligonucleotide and 1 µg (lanes 1, 3, 5 and 7) or 3 µg (lanes 2, 4, 6 and 8) protein extract.
Figure 4
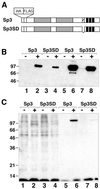
Acetylation of Sp3 in vivo. Stable SL2 transfectants containing expression constructs for epitope-tagged Sp3 or Sp3SD were incubated with [3H]acetate (1 mCi/ml) for 1 h. Nuclear extracts were prepared and subjected to immunoprecipitation with an αFLAG antibody. (A) Schematic representation of Sp3 and the mutant Sp3SD lacking a 13 amino acid stretch. (B) Nuclear extracts prepared from SL2 cells prior to (lanes 1–4) and after (lanes 5–8) immunoprecipitation with an αFLAG antibody were subjected to western blot analyses with an αHA antiserum and visualized by chemoluminescence detection. (–) and (+) indicate the absence and presence of copper sulfate (induction of Sp3 protein). (C) Autoradiogram showing [3H]acetate-labeled proteins of the same fractions as in (B).
Figure 5
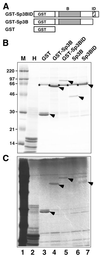
In vitro acetylation of recombinant Sp3 fusion proteins. (A) Schematic representation of GST fusion proteins used as substrates. Sp3B and Sp3BID were obtained by thrombin cleavage of the appropriate GST fusion proteins. GST, GST–Sp3BID, GST–Sp3B, Sp3B, Sp3BID and histones were incubated with GST–CBP-HAT in the presence of [14C]acetate and analyzed by SDS–PAGE and autoradiography. (B) Coomassie stained SDS–PAGE gel of fractionated proteins subjected to the in vitro acetylation reaction. Lane M is the marker lane and lane H contains histones. The stars indicate BSA that was included in the acetylation reactions. The arrows point to the GST, GST–Sp3BID, GST–Sp3B, Sp3B and Sp3BID proteins. (C) Autoradiogram of the gel shown in (B). The marker lane (M) contains 14C-labeled proteins. The arrows point to the acetylated GST, GST–Sp3BID, GST–Sp3B and Sp3BID proteins.
Similar articles
- An inhibitor domain in Sp3 regulates its glutamine-rich activation domains.
Dennig J, Beato M, Suske G. Dennig J, et al. EMBO J. 1996 Oct 15;15(20):5659-67. EMBO J. 1996. PMID: 8896459 Free PMC article. - Identification of the promoter of human transcription factor Sp3 and evidence of the role of factors Sp1 and Sp3 in the expression of Sp3 protein.
Lou Z, Maher VM, McCormick JJ. Lou Z, et al. Gene. 2005 May 23;351:51-9. doi: 10.1016/j.gene.2005.02.007. Epub 2005 Apr 25. Gene. 2005. PMID: 15857802 - Estrogen regulation of trefoil factor 1 expression by estrogen receptor alpha and Sp proteins.
Sun JM, Spencer VA, Li L, Yu Chen H, Yu J, Davie JR. Sun JM, et al. Exp Cell Res. 2005 Jan 1;302(1):96-107. doi: 10.1016/j.yexcr.2004.08.015. Exp Cell Res. 2005. PMID: 15541729 - Sp1 control of gene expression in myeloid cells.
Resendes KK, Rosmarin AG. Resendes KK, et al. Crit Rev Eukaryot Gene Expr. 2004;14(3):171-81. doi: 10.1615/critreveukaryotgeneexpr.v14.i3.20. Crit Rev Eukaryot Gene Expr. 2004. PMID: 15248814 Review.
Cited by
- Transcription factor Sp3 is silenced through SUMO modification by PIAS1.
Sapetschnig A, Rischitor G, Braun H, Doll A, Schergaut M, Melchior F, Suske G. Sapetschnig A, et al. EMBO J. 2002 Oct 1;21(19):5206-15. doi: 10.1093/emboj/cdf510. EMBO J. 2002. PMID: 12356736 Free PMC article. - Modulation of Hepatic Insulin and Glucagon Signaling by Nutritional Factors in Broiler Chicken.
Petrilla J, Mátis G, Mackei M, Kulcsár A, Sebők C, Papp M, Gálfi P, Fébel H, Huber K, Neogrády Z. Petrilla J, et al. Vet Sci. 2022 Feb 25;9(3):103. doi: 10.3390/vetsci9030103. Vet Sci. 2022. PMID: 35324832 Free PMC article. - Molecular cloning, epigenetic regulation, and functional characterization of Prkd1 gene promoter in dopaminergic cell culture models of Parkinson's disease.
Ay M, Jin H, Harischandra DS, Asaithambi A, Kanthasamy A, Anantharam V, Kanthasamy AG. Ay M, et al. J Neurochem. 2015 Oct;135(2):402-15. doi: 10.1111/jnc.13261. Epub 2015 Aug 28. J Neurochem. 2015. PMID: 26230914 Free PMC article. - Posttranslational modifications in diabetes: Mechanisms and functions.
Hu A, Zou H, Chen B, Zhong J. Hu A, et al. Rev Endocr Metab Disord. 2022 Oct;23(5):1011-1033. doi: 10.1007/s11154-022-09740-x. Epub 2022 Jun 13. Rev Endocr Metab Disord. 2022. PMID: 35697961 Review. - Functional characterization of the human translocator protein (18kDa) gene promoter in human breast cancer cell lines.
Batarseh A, Barlow KD, Martinez-Arguelles DB, Papadopoulos V. Batarseh A, et al. Biochim Biophys Acta. 2012 Jan;1819(1):38-56. doi: 10.1016/j.bbagrm.2011.09.001. Epub 2011 Sep 18. Biochim Biophys Acta. 2012. PMID: 21958735 Free PMC article.
References
- Suske G. (1999) The Sp-family of transcription factors. Gene, 238, 291–300. - PubMed
- Marin M., Karis,A., Visser,P., Grosveld,F. and Philipsen,S. (1997) Transcription factor Sp1 is essential for early development but dispensable for cell growth and differentiation. Cell, 89, 619–628. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous