Flexibility and packing in proteins - PubMed (original) (raw)
Flexibility and packing in proteins
Bertil Halle. Proc Natl Acad Sci U S A. 2002.
Abstract
Structural flexibility is an essential attribute, without which few proteins could carry out their biological functions. Much information about protein flexibility has come from x-ray crystallography, in the form of atomic mean-square displacements (AMSDs) or B factors. Profiles showing the AMSD variation along the polypeptide chain are usually interpreted in dynamical terms but are ultimately governed by the local features of a highly complex energy landscape. Here, we bypass this complexity by showing that the AMSD profile is essentially determined by spatial variations in local packing density. On the basis of elementary statistical mechanics and generic features of atomic distributions in proteins, we predict a direct inverse proportionality between the AMSD and the contact density, i.e., the number of noncovalent neighbor atoms within a local region of approximately 1.5 nm(3) volume. Testing this local density model against a set of high-quality crystal structures of 38 nonhomologous proteins, we find that it accurately and consistently reproduces the prominent peaks in the AMSD profile and even captures minor features, such as the periodic AMSD variation within alpha helices. The predicted rigidifying effect of crystal contacts also agrees with experimental data. With regard to accuracy and computational efficiency, the model is clearly superior to its predecessors. The quantitative link between flexibility and packing density found here implies that AMSDs provide little independent information beyond that contained in the mean atomic coordinates.
Figures
Figure 1
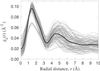
Radial distribution of nonhydrogen atoms around each α carbon in parvalbumin (2
pvb
), computed from Eq. 6. The thick black curve is the average of the 107 gray curves.
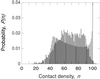
Figure 2
Normalized contact density distribution (binned to integral n values) for all 6,231 α carbons in the set of 38 proteins (black) and for the uniform-sphere model described in the text (gray). For the latter case, the n = 100 bar has been truncated at about 25% of its real height, P(100) = 0.196.
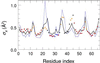
Figure 3
Experimental (dots) and calculated (curve) AMSD profiles for the α carbons in S. marcescens endonuclease (1
ql
0). Predicted AMSDs are based on contact densities including all nonhydrogen protein atoms in the crystal. Experimental points are color coded according to secondary structure: α helix (blue), β strand (red), and turn (orange).
Figure 4
Experimental (dots) and calculated (curves) AMSD profiles for the α carbons in B. caldolyticus cold-shock protein (1
c
9
o
). Predicted AMSDs are based on contact densities including all nonhydrogen protein atoms in the crystal (thick black curve) or only atoms in the same protein molecule as the reference α carbons (thin blue curve). Experimental points are color coded according to secondary structure: α helix (blue), β strand (red), and turn (orange).
Figure 5
AMSD profile for all nonhydrogen atoms in the Kunitz-type domain (C5) from the α-3 chain of human type VI collagen (2
knt
). Circles represent experimental backbone (filled) and side-chain (open) AMSDs, and curves represent predicted AMSDs on the basis of contact densities including all nonhydrogen protein atoms in the crystal (thick black curve, Δ = 0.63, ρ = 0.72) or only atoms in the same protein molecule as the reference atoms (thin blue curve, Δ = 0.64, ρ = 0.76). Experimental points for atoms in disulfide Cys residues are colored orange.
Similar articles
- Sequence Conservation, Radial Distance and Packing Density in Spherical Viral Capsids.
Chang CM, Huang YW, Lee CW, Huang TT, Shih CS, Hwang JK. Chang CM, et al. PLoS One. 2015 Jul 1;10(7):e0132234. doi: 10.1371/journal.pone.0132234. eCollection 2015. PLoS One. 2015. PMID: 26132081 Free PMC article. - A large data set comparison of protein structures determined by crystallography and NMR: statistical test for structural differences and the effect of crystal packing.
Andrec M, Snyder DA, Zhou Z, Young J, Montelione GT, Levy RM. Andrec M, et al. Proteins. 2007 Nov 15;69(3):449-65. doi: 10.1002/prot.21507. Proteins. 2007. PMID: 17623851 - Protein-protein crystal-packing contacts.
Carugo O, Argos P. Carugo O, et al. Protein Sci. 1997 Oct;6(10):2261-3. doi: 10.1002/pro.5560061021. Protein Sci. 1997. PMID: 9336849 Free PMC article. Review. - The limit of accuracy of protein modeling: influence of crystal packing on protein structure.
Eyal E, Gerzon S, Potapov V, Edelman M, Sobolev V. Eyal E, et al. J Mol Biol. 2005 Aug 12;351(2):431-42. doi: 10.1016/j.jmb.2005.05.066. J Mol Biol. 2005. PMID: 16005885 - Measuring the hydrogen/deuterium exchange of proteins at high spatial resolution by mass spectrometry: overcoming gas-phase hydrogen/deuterium scrambling.
Rand KD, Zehl M, Jørgensen TJ. Rand KD, et al. Acc Chem Res. 2014 Oct 21;47(10):3018-27. doi: 10.1021/ar500194w. Epub 2014 Aug 29. Acc Chem Res. 2014. PMID: 25171396 Review.
Cited by
- Improved prediction of site-rates from structure with averaging across homologs.
Norn C, Oliveira F, André I. Norn C, et al. Protein Sci. 2024 Jul;33(7):e5086. doi: 10.1002/pro.5086. Protein Sci. 2024. PMID: 38923241 Free PMC article. - Deciphering the preference and predicting the viability of circular permutations in proteins.
Lo WC, Dai T, Liu YY, Wang LF, Hwang JK, Lyu PC. Lo WC, et al. PLoS One. 2012;7(2):e31791. doi: 10.1371/journal.pone.0031791. Epub 2012 Feb 16. PLoS One. 2012. PMID: 22359629 Free PMC article. - Protein structural variation in computational models and crystallographic data.
Kondrashov DA, Van Wynsberghe AW, Bannen RM, Cui Q, Phillips GN Jr. Kondrashov DA, et al. Structure. 2007 Feb;15(2):169-77. doi: 10.1016/j.str.2006.12.006. Structure. 2007. PMID: 17292835 Free PMC article. - Protein mechanics: a route from structure to function.
Lavery R, Sacquin-Mora S. Lavery R, et al. J Biosci. 2007 Aug;32(5):891-8. doi: 10.1007/s12038-007-0089-x. J Biosci. 2007. PMID: 17914231 Review. - Protein hydration dynamics in solution: a critical survey.
Halle B. Halle B. Philos Trans R Soc Lond B Biol Sci. 2004 Aug 29;359(1448):1207-23; discussion 1223-4, 1323-8. doi: 10.1098/rstb.2004.1499. Philos Trans R Soc Lond B Biol Sci. 2004. PMID: 15306377 Free PMC article. Review.
References
- Willis B T M, Pryor A W. Thermal Vibrations in Crystallography. Cambridge, U.K.: Cambridge Univ. Press; 1975.
- Trueblood K N, Bürgi H-B, Burzlaff H, Dunitz J D, Gramaccioli C M, Schulz H H, Shmueli U, Abrahams S C. Acta Crystallogr A. 1996;52:770–781.
- Ringe D, Petsko G A. Prog Biophys Mol Biol. 1985;45:197–235. - PubMed
- Frauenfelder H, Petsko G A, Tsernoglou D. Nature (London) 1979;280:558–563. - PubMed
- Artymiuk P J, Blake C C F, Grace D E P, Oatley S J, Phillips D C, Sternberg M J E. Nature (London) 1979;280:563–568. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources