Human Elongator facilitates RNA polymerase II transcription through chromatin - PubMed (original) (raw)
Human Elongator facilitates RNA polymerase II transcription through chromatin
Jae-Hyun Kim et al. Proc Natl Acad Sci U S A. 2002.
Abstract
A human Elongator complex was purified from HeLa cells and found to be composed of three polypeptides. Human Elongator contains histone acetyltransferase activity with specificity to histone H3 and, to a much lesser extent, to histone H4. Although many reports have suggested a role for the yeast Elongator in transcription elongation through chromatin templates, no direct evidence supporting this function exists. In the present study, we demonstrate that the human Elongator facilitates transcription by RNA polymerase II in a chromatin- and acetyl-CoA-dependent manner. The complex was found to directly interact with RNA polymerase II but failed to interact with other factors that facilitated RNA polymerase II to traverse through nucleosomes. From our results, we postulate that different mechanisms operate to ensure efficient transcription by RNA polymerase II on chromatin templates.
Figures
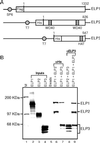
Figure 1
Purification of a hElongator. (A) Schematic representation of the steps used to purify a hElongator. (B) Fractions derived from each step of purification and containing approximately equal amounts of protein were resolved by electrophoresis on an SDS/4–15% polyacrylamide gel and stained with silver. (C Upper) Silver staining of an SDS/10% polyacrylamide gel containing aliquots of fractions derived from the Superose-6 gel filtration column. The subunits of core Elongator are indicated on the left, and the elution from the gel filtration column of protein markers is indicated on the top. (Lower) Western blot of the fractions derived from the gel filtration column analyzed by using antibodies specific for Elp1 and Elp3. (D) Human Elp3 is an HAT. (Upper) Schematic representation of human Elp3-conserved motifs D, A, and B of the GNAT superfamily (34) is indicated. The Q(R)XXGXG motif (central part of GNAT motif A) critical for acetyl-CoA recognition (35) is conserved in the human Elp3 subunit. Numbers on the top denote the amino acid sequence of hElp3. (Lower) HAT assay was performed by using the purified hElongator (20 and 100 ng of the gel filtration pool). P/CAF used as control is indicated. [3H]Acetylated core histones were analyzed by SDS/15% PAGE followed by fluorography.
Figure 2
Subunit interaction of the core human Elongator. (A) Schematic representation of N-terminally tagged human Elp1, Elp2, and Elp3. Numbers on the top indicate the N- and C-terminal amino acids of each construct. The promoters used to transcribe the DNA in vitro are indicated. (B) Elp1 interacts with Elp2 and with Elp3. In vitro translated N-terminally His-tagged Elp1 was incubated with in vitro translated N-terminally flag-tagged Elp2 and/or Elp3. The complex was immunoprecipitated with antibodies against the His-tag or Elp3 as described in Materials and Methods. Immunoprecipitates were separated by SDS/10% PAGE and analyzed by autoradiography.
Figure 3
Subcellular localization of human core-Elongator subunits. HeLa cells were fixed and stained with various fluorescent-conjugated antibodies or 0.01 mg/ml 4′,6-diamidino-2-phenylindole (DAPI) as indicated.
Figure 4
Human core Elongator interacts with RNAPII (Pol II) in vitro. Elongator (100 ng) and RNAPII (300 ng) were incubated and then fractionated through a Superose-6 column as indicated in Materials and Methods. In parallel experiments, individual factors (top of each gel) were also fractionated on the column. The bottom of each gel denotes the fractions derived from the Elongator–RNAPII fractionation. The indicated fractions were blotted and probed with anti-Elp1, anti-Elp3, and anti-CTD as indicated. Elution positions of size markers are indicated on the top.
Figure 5
Human Elongator allows transcription from chromatin templates. (A) Immunoblot analysis of untreated HeLa NE and Elongator-depleted HeLa NE. Equal amounts (30 μg) of NE and Elongator-depleted NE were subjected to SDS/4–15% polyacrylamide gradient gel electrophoresis, transferred to nitrocellulose, and analyzed with antibodies to the proteins indicated. (B) Transcription from naked DNA and chromatin templates. Transcription was performed by using either normal NE or Elongator-depleted NE (NE[ΔELP]). Transcription reaction mixtures (40 μl) contained 80 μg of NE proteins and freshly assembled chromatin or histone-free DNA (100 ng) with (20, 40, and 80 ng) or without Elongator and acetyl-CoA (1.5 μM), as indicated. In addition, reactions were supplemented with Gal4-VP16. Reactions were incubated at 30°C for 30 min or as indicated. Products of the transcription reactions were separated by electrophoresis on a 6% sequencing gel.
Similar articles
- Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo.
Winkler GS, Kristjuhan A, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Winkler GS, et al. Proc Natl Acad Sci U S A. 2002 Mar 19;99(6):3517-22. doi: 10.1073/pnas.022042899. Proc Natl Acad Sci U S A. 2002. PMID: 11904415 Free PMC article. - Purification and characterization of the human elongator complex.
Hawkes NA, Otero G, Winkler GS, Marshall N, Dahmus ME, Krappmann D, Scheidereit C, Thomas CL, Schiavo G, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Hawkes NA, et al. J Biol Chem. 2002 Jan 25;277(4):3047-52. doi: 10.1074/jbc.M110445200. Epub 2001 Nov 19. J Biol Chem. 2002. PMID: 11714725 - Purified histone acetyltransferase complexes stimulate HIV-1 transcription from preassembled nucleosomal arrays.
Steger DJ, Eberharter A, John S, Grant PA, Workman JL. Steger DJ, et al. Proc Natl Acad Sci U S A. 1998 Oct 27;95(22):12924-9. doi: 10.1073/pnas.95.22.12924. Proc Natl Acad Sci U S A. 1998. PMID: 9789016 Free PMC article. - TBP-associated factors in the PCAF histone acetylase complex.
Kotani T, Zhang X, Schiltz RL, Ogryzko VV, Howard T, Swanson MJ, Vassilev A, Zhang H, Yamauchi J, Howard BH, Qin J, Nakatani Y. Kotani T, et al. Cold Spring Harb Symp Quant Biol. 1998;63:493-9. doi: 10.1101/sqb.1998.63.493. Cold Spring Harb Symp Quant Biol. 1998. PMID: 10384313 Review. No abstract available. - Elongator complex: how many roles does it play?
Svejstrup JQ. Svejstrup JQ. Curr Opin Cell Biol. 2007 Jun;19(3):331-6. doi: 10.1016/j.ceb.2007.04.005. Epub 2007 Apr 26. Curr Opin Cell Biol. 2007. PMID: 17466506 Review.
Cited by
- Transcriptome analysis in a humanized mouse model of familial dysautonomia reveals tissue-specific gene expression disruption in the peripheral nervous system.
Harripaul R, Morini E, Salani M, Logan E, Kirchner E, Bolduc J, Chekuri A, Currall B, Yadav R, Erdin S, Talkowski ME, Gao D, Slaugenhaupt S. Harripaul R, et al. Sci Rep. 2024 Jan 4;14(1):570. doi: 10.1038/s41598-023-51137-6. Sci Rep. 2024. PMID: 38177237 Free PMC article. - The life and times of a tRNA.
Phizicky EM, Hopper AK. Phizicky EM, et al. RNA. 2023 Jul;29(7):898-957. doi: 10.1261/rna.079620.123. Epub 2023 Apr 13. RNA. 2023. PMID: 37055150 Free PMC article. Review. - Development of an oral treatment that rescues gait ataxia and retinal degeneration in a phenotypic mouse model of familial dysautonomia.
Morini E, Chekuri A, Logan EM, Bolduc JM, Kirchner EG, Salani M, Krauson AJ, Narasimhan J, Gabbeta V, Grover S, Dakka A, Mollin A, Jung SP, Zhao X, Zhang N, Zhang S, Arnold M, Woll MG, Naryshkin NA, Weetall M, Slaugenhaupt SA. Morini E, et al. Am J Hum Genet. 2023 Mar 2;110(3):531-547. doi: 10.1016/j.ajhg.2023.01.019. Epub 2023 Feb 20. Am J Hum Genet. 2023. PMID: 36809767 Free PMC article. - HITS-CLIP analysis of human ALKBH8 reveals interactions with fully processed substrate tRNAs and with specific noncoding RNAs.
Cavallin I, Bartosovic M, Skalicky T, Rengaraj P, Demko M, Schmidt-Dengler MC, Drino A, Helm M, Vanacova S. Cavallin I, et al. RNA. 2022 Dec;28(12):1568-1581. doi: 10.1261/rna.079421.122. Epub 2022 Oct 3. RNA. 2022. PMID: 36192131 Free PMC article. - Rescue of a familial dysautonomia mouse model by AAV9-Exon-specific U1 snRNA.
Romano G, Riccardi F, Bussani E, Vodret S, Licastro D, Ragone I, Ronzitti G, Morini E, Slaugenhaupt SA, Pagani F. Romano G, et al. Am J Hum Genet. 2022 Aug 4;109(8):1534-1548. doi: 10.1016/j.ajhg.2022.07.004. Epub 2022 Jul 28. Am J Hum Genet. 2022. PMID: 35905737 Free PMC article.
References
- Orphanides G, Reinberg D. Nature (London) 2000;407:471–475. - PubMed
- Renner D B, Yamaguchi Y, Wada T, Handa H, Price D H. J Biol Chem. 2001;276:42601–42609. - PubMed
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. Cell. 1999;97:41–51. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases