Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins - PubMed (original) (raw)
Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins
Malik Lutzmann et al. EMBO J. 2002.
Abstract
Now that it is likely that all yeast nucleoporins are known, one of the ultimate goals is the in vitro assembly of the entire nuclear pore complex from its approximately 30 individual components. Here, we report the reconstitution of seven proteins (Nup133p, Nup145p-C, Nup120p, Nup85p, Nup84p, Seh1p and Sec13p) into a heptameric 0.5 MDa nuclear pore subcomplex. We found that double plasmid transformation combined with bi-cistronic mRNA translation allow the expression and assembly of distinct subcomplexes of up to five nucleoporins in a single Escherichia coli cell. During the sequential reconstitution of the Nup84p complex, smaller assembly intermediates can be isolated, which exhibit modular structures determined by electron microscopy that finally make up the whole Y-shaped Nup84p complex. Importantly, a seventh subunit, Nup133p, was incorporated into the complex through its interaction with Nup84p, thereby elongating one arm of the Y-shaped assembly to an approximately 40 nm long stalk. Taken together, our data document that the Nup84p-Nup133p complex self-assembles in a modular concept from distinct smaller nucleoporin construction sets.
Figures
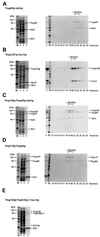
Fig. 1. Expression of nucleoporins of the Nup84p complex in E.coli. BL21 cells were transformed with plasmids harboring the indicated nucleoporin genes. Cells were grown under non-induced (–) and IPTG-induced (+) conditions. Whole-cell lysates were prepared and analyzed by SDS–PAGE and Coomassie Blue staining. The positions of the induced nucleoporins are indicated by asterisks. In (A), single nucleoporins were expressed, whereas in (B) pairs of nucleoporins were expressed from bi-cistronic constructs. Also shown is a 10 kDa molecular weight ladder.
Fig. 2. Isolation of heterodimeric and heterotrimeric nucleoporin complexes. Purification of (A) the Nup85p–Seh1p complex, (B) the Nup145p-C–Sec13p complex, (C) the Nup120p–Nup85p–Seh1p complex, (D) the Nup133p–Nup84p complex and (E) the Nup120p–Nup145p-C–Sec13p complex. Note that the band below Nup145p-C is a degradation product of this protein as shown by western blot analysis (data not shown). Left panels (A–E): Coomassie-stained SDS–PAGE with a protein standard (M), the soluble E.coli lysate after high speed centrifugation (lane 1), the flow through after incubation with glutathione beads (lane 2) and the TEV eluate (lane 3). Right panels (A–D): the TEV eluates containing the indicated complexes were applied on an FPLC Superdex 200 column. Fractions 13–25 from the column were analyzed by SDS–PAGE and Coomassie Blue staining. Also shown is the TEV eluate input fraction (L) and a protein standard (M). The Superdex 200 column was calibrated with several molecular weight marker proteins of 670, 158, 44 and 17 kDa (Bio-Rad).
Fig. 3. Purification of the reconstituted Nup84p complex with or without Nup133p. The upper gels show the in vitro assembled Nup84p complex without Nup133p (A) and with bound Nup133p (B) before Superose 6 gel filtration chromatography (input fraction = L), a protein standard (M) and fractions 16–28 from the first Superose 6 column run. The lower gels are the result of a second Superose gel filtration of the pooled fractions 21–24 (L). Note that the additional band at ∼65 kDa is a degradation product of Nup145p-C (revealed by western blot; data not shown). The Superose 6 column was calibrated with molecular weight marker proteins of 670, 158, 44 and 17 kDa (Bio-Rad). A typical calibration profile is shown at the bottom of (A).
Fig. 3. Purification of the reconstituted Nup84p complex with or without Nup133p. The upper gels show the in vitro assembled Nup84p complex without Nup133p (A) and with bound Nup133p (B) before Superose 6 gel filtration chromatography (input fraction = L), a protein standard (M) and fractions 16–28 from the first Superose 6 column run. The lower gels are the result of a second Superose gel filtration of the pooled fractions 21–24 (L). Note that the additional band at ∼65 kDa is a degradation product of Nup145p-C (revealed by western blot; data not shown). The Superose 6 column was calibrated with molecular weight marker proteins of 670, 158, 44 and 17 kDa (Bio-Rad). A typical calibration profile is shown at the bottom of (A).
Fig. 4. The reconstituted intermediate heteromeric complexes display modular structures visualized by EM analysis after purification and negative staining. Shown is a Coomassie-stained SDS–polyacrylamide gel of the purified protein or complex (left panels), an overview electron micrograph of the negatively stained preparation (middle panels) and a gallery of selected particles (right panels). (A) Nup84p; (B) Nup85p–Seh1p complex; (C) Nup145p-C–Sec13p complex; (D) Nup120p–Nup85p–Seh1p complex; (E) Nup133p–Nup84p complex; (F) Nup133p–Nup84p–Nup145p-C–Sec13p complex.
Fig. 4. The reconstituted intermediate heteromeric complexes display modular structures visualized by EM analysis after purification and negative staining. Shown is a Coomassie-stained SDS–polyacrylamide gel of the purified protein or complex (left panels), an overview electron micrograph of the negatively stained preparation (middle panels) and a gallery of selected particles (right panels). (A) Nup84p; (B) Nup85p–Seh1p complex; (C) Nup145p-C–Sec13p complex; (D) Nup120p–Nup85p–Seh1p complex; (E) Nup133p–Nup84p complex; (F) Nup133p–Nup84p–Nup145p-C–Sec13p complex.
Fig. 5. The reconstituted Nup84p complex displays a Y-shaped structure, in which one arm, the ‘stalk’ of the ‘Y’, is extended by Nup133p. Shown is the complex purified by SDS–PAGE with Coomassie Blue staining (left panels), an overview electron micrograph of the negatively stained preparation (middle panels) and a gallery of selected particles (right panels). (A) In vitro reconstituted hexameric Nup84p complex. (B) In vitro reconstituted pentameric Nup84p complex lacking the Nup84p subunit. (C) In vitro reconstituted heptameric Nup84p complex with bound Nup133p.
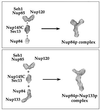
Fig. 6. Structural model of the Nup84p–Nup133p complex. Representative single particles of the different subcomplexes and the whole Nup84p complex with or without Nup133p (see Figures 4 and 5) were taken from scanned electron micrographs using Adobe Photoshop. After magnification by the same factor, the pictures were blurred to overcome the pixelated appearance and arranged as depicted. The modular assembly of the Nup84p complex from the different subcomplexes and the elongation of its stalk by adding Nup133p to Nup84p are apparent. Note that the depicted EM pictures are processed extensively and thus should be considered only as models. The labels of the binary complexes (e.g. Nup145p-C–Sec13p, Nup85p–Seh1p) do not imply exactly where each subunit within the heterodimer is located.
Similar articles
- Structure and assembly of the Nup84p complex.
Siniossoglou S, Lutzmann M, Santos-Rosa H, Leonard K, Mueller S, Aebi U, Hurt E. Siniossoglou S, et al. J Cell Biol. 2000 Apr 3;149(1):41-54. doi: 10.1083/jcb.149.1.41. J Cell Biol. 2000. PMID: 10747086 Free PMC article. - A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores.
Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt EC. Siniossoglou S, et al. Cell. 1996 Jan 26;84(2):265-75. doi: 10.1016/s0092-8674(00)80981-2. Cell. 1996. PMID: 8565072 - Two functionally distinct domains generated by in vivo cleavage of Nup145p: a novel biogenesis pathway for nucleoporins.
Teixeira MT, Siniossoglou S, Podtelejnikov S, Bénichou JC, Mann M, Dujon B, Hurt E, Fabre E. Teixeira MT, et al. EMBO J. 1997 Aug 15;16(16):5086-97. doi: 10.1093/emboj/16.16.5086. EMBO J. 1997. PMID: 9305650 Free PMC article. - Structural analysis of the nuclear pore complex by integrated approaches.
Elad N, Maimon T, Frenkiel-Krispin D, Lim RY, Medalia O. Elad N, et al. Curr Opin Struct Biol. 2009 Apr;19(2):226-32. doi: 10.1016/j.sbi.2009.02.009. Epub 2009 Mar 25. Curr Opin Struct Biol. 2009. PMID: 19327984 Review. - Gene regulation by nucleoporins and links to cancer.
Köhler A, Hurt E. Köhler A, et al. Mol Cell. 2010 Apr 9;38(1):6-15. doi: 10.1016/j.molcel.2010.01.040. Mol Cell. 2010. PMID: 20385085 Review.
Cited by
- The yeast nuclear pore complex and transport through it.
Aitchison JD, Rout MP. Aitchison JD, et al. Genetics. 2012 Mar;190(3):855-83. doi: 10.1534/genetics.111.127803. Genetics. 2012. PMID: 22419078 Free PMC article. - Analysis of the initiation of nuclear pore assembly by ectopically targeting nucleoporins to chromatin.
Schwartz M, Travesa A, Martell SW, Forbes DJ. Schwartz M, et al. Nucleus. 2015;6(1):40-54. doi: 10.1080/19491034.2015.1004260. Nucleus. 2015. PMID: 25602437 Free PMC article. - A jumbo problem: mapping the structure and functions of the nuclear pore complex.
Fernandez-Martinez J, Rout MP. Fernandez-Martinez J, et al. Curr Opin Cell Biol. 2012 Feb;24(1):92-9. doi: 10.1016/j.ceb.2011.12.013. Epub 2012 Feb 8. Curr Opin Cell Biol. 2012. PMID: 22321828 Free PMC article. Review. - Resolving the composition of protein complexes using a MALDI LTQ Orbitrap.
Luo Y, Li T, Yu F, Kramer T, Cristea IM. Luo Y, et al. J Am Soc Mass Spectrom. 2010 Jan;21(1):34-46. doi: 10.1016/j.jasms.2009.08.026. Epub 2009 Sep 17. J Am Soc Mass Spectrom. 2010. PMID: 19822444 Free PMC article. - A versatile interaction platform on the Mex67-Mtr2 receptor creates an overlap between mRNA and ribosome export.
Yao W, Lutzmann M, Hurt E. Yao W, et al. EMBO J. 2008 Jan 9;27(1):6-16. doi: 10.1038/sj.emboj.7601947. Epub 2007 Nov 29. EMBO J. 2008. PMID: 18046452 Free PMC article.
References
- Allen N.P., Huang,L., Burlingame,A. and Rexach,M.F. (2001) Proteomic analysis of nucleoporin interacting proteins. J. Biol. Chem., 276, 29268–29274. - PubMed
- Allen T.D., Cronshaw,J.M., Bagley,S., Kiseleva,E. and Goldberg,M.W. (2000) The nuclear pore complex: mediator of translocation between nucleus and cytoplasm. J. Cell Sci., 113, 1651–1659. - PubMed
- Bagley S., Goldberg,M.W., Cronshaw,J.M., Rutherford,S.A. and Allen,T.D. (2000) The nuclear pore complex. J. Cell Sci., 113, 3885–3886. - PubMed
- Bayliss R., Littlewood,T. and Stewart,M. (2000) Structural basis for the interaction between FxFG nucleoporin repeats and importin-β in nuclear trafficking. Cell, 102, 99–108. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases